Create Cluster Heatmap¶
-
showClusterHeatmap()
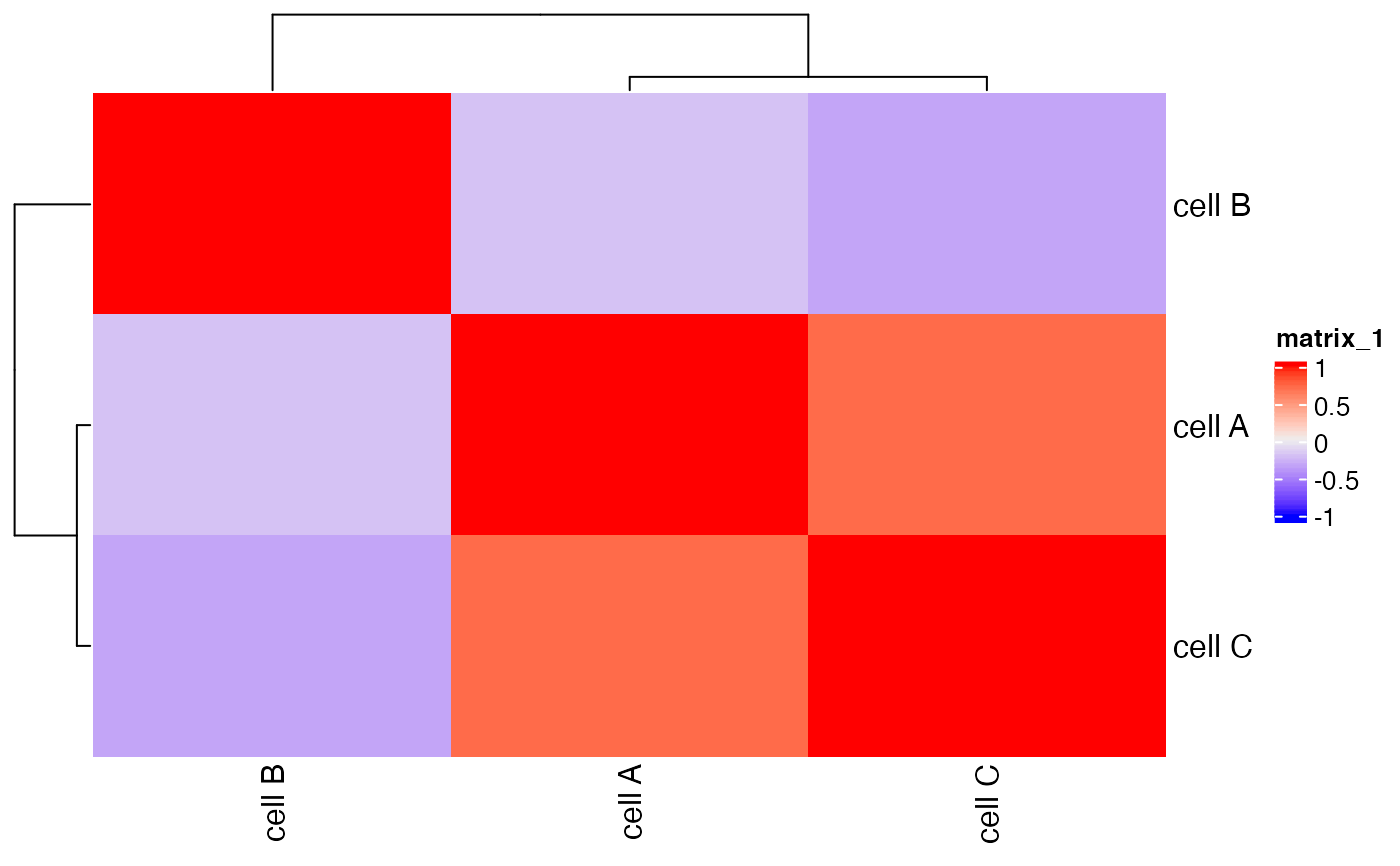
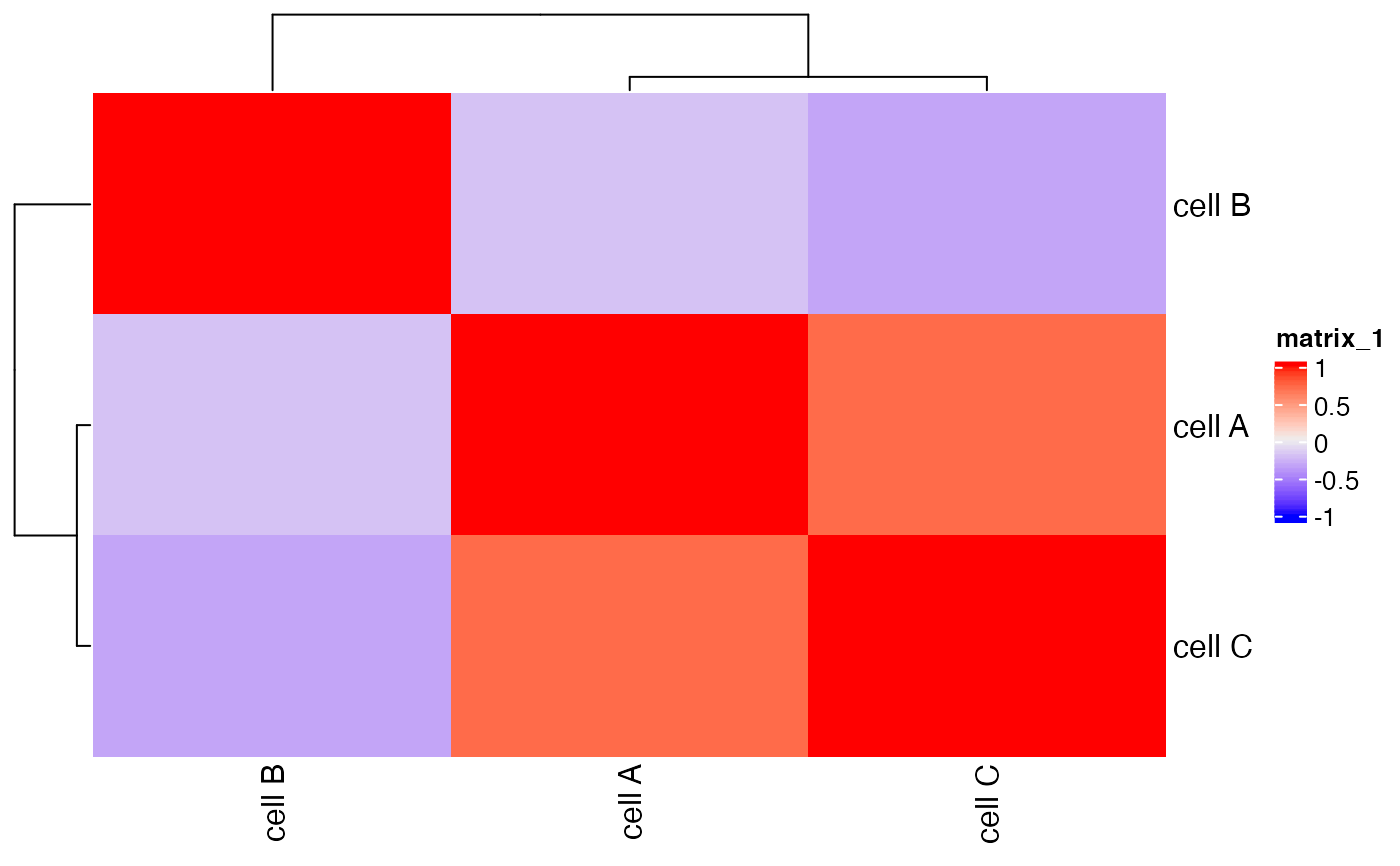
Creates heatmap based on identified clusters.
showClusterHeatmap(
gobject,
expression_values = c("normalized", "scaled", "custom"),
genes = "all",
cluster_column,
cor = c("pearson", "spearman"),
distance = "ward.D",
show_plot = NA,
return_plot = NA,
save_plot = NA,
save_param = list(),
default_save_name = "showClusterHeatmap",
...
)
Arguments¶
gobject |
giotto object |
expression_values |
expression values to use |
genes |
vector of genes to use, default to ‘all’ |
cluster_column |
name of column to use for clusters |
cor |
correlation score to calculate distance |
distance |
distance method to use for hierarchical clustering |
show_plot |
show plot |
return_plot |
return ggplot object |
save_plot |
directly save the plot [boolean] |
save_param |
list of saving parameters, see showSaveParameters. |
default_save_name |
default save name for saving, don’t change, change save_name in save_param |
… |
additional parameters for the Heatmap function from ComplexHeatmap |
Value¶
A ggplot value.
Details¶
Correlation heatmap of selected clusters.
Examples¶
data(mini_giotto_single_cell)
# cell metadata
cell_metadata = pDataDT(mini_giotto_single_cell)
# create heatmap
showClusterHeatmap(mini_giotto_single_cell,
cluster_column = 'cell_types')