Visualize Cells By Spatial And Dimension Coordinates in 2D¶
-
spatDimCellPlot()
Visualize numerical features of cells according to spatial AND dimension reduction coordinates in 2D.
spatDimCellPlot(...)
Arguments¶
… |
Arguments passed on to spatDimCellPlot2D() |
gobject |
giotto object |
show_image |
show a tissue background image |
gimage |
a giotto image |
image_name |
name of a giotto image |
plot_alignment |
direction to align plot |
spat_enr_names |
names of spatial enrichment results to include |
cell_annotation_values |
numeric cell annotation columns |
dim_reduction_to_use |
dimension reduction to use |
dim_reduction_name |
dimension reduction name |
dim1_to_use |
dimension to use on x-axis |
dim2_to_use |
dimension to use on y-axis |
sdimx |
= spatial dimension to use on x-axis |
sdimy |
= spatial dimension to use on y-axis |
cell_color_gradient |
vector with 3 colors for numeric data |
gradient_midpoint |
midpoint for color gradient |
gradient_limits |
vector with lower and upper limits |
select_cell_groups |
select subset of cells/clusters based on cell_color parameter |
select_cells |
select subset of cells based on cell IDs |
dim_point_shape |
dim reduction points with border or not (border or no_border) |
dim_point_size |
size of points in dim. reduction space |
dim_point_alpha |
transparency of dim. reduction points |
dim_point_border_col |
border color of points in dim. reduction space |
dim_point_border_stroke |
border stroke of points in dim. reduction space |
spat_point_shape |
shape of points (border, no_border or voronoi) |
spat_point_size |
size of spatial points |
spat_point_alpha |
transparency of spatial points |
spat_point_border_col |
border color of spatial points |
spat_point_border_stroke |
border stroke of spatial points |
dim_show_cluster_center |
show the center of each cluster |
dim_show_center_label |
provide a label for each cluster |
dim_center_point_size |
size of the center point |
dim_center_point_border_col |
border color of center point |
dim_center_point_border_stroke |
stroke size of center point |
dim_label_size |
size of the center label |
dim_label_fontface |
font of the center label |
spat_show_cluster_center |
show the center of each cluster |
spat_show_center_label |
provide a label for each cluster |
spat_center_point_size |
size of the spatial center points |
spat_center_point_border_col |
border color of the spatial center points |
spat_center_point_border_stroke |
stroke size of the spatial center points |
spat_label_size |
size of the center label |
spat_label_fontface |
font of the center label |
show_NN_network |
show underlying NN network |
nn_network_to_use |
type of NN network to use (kNN vs sNN) |
nn_network_name |
name of NN network to use, if show_NN_network = TRUE |
dim_edge_alpha |
column to use for alpha of the edges |
spat_show_network |
show spatial network |
spatial_network_name |
name of spatial network to use |
spat_network_color |
color of spatial network |
spat_network_alpha |
alpha of spatial network |
spat_show_grid |
show spatial grid |
spatial_grid_name |
name of spatial grid to use |
spat_grid_color |
color of spatial grid |
show_other_cells |
display not selected cells |
other_cell_color |
color of not selected cells |
dim_other_point_size |
size of not selected dim cells |
spat_other_point_size |
size of not selected spat cells |
spat_other_cells_alpha |
alpha of not selected spat cells |
coord_fix_ratio |
ratio for coordinates |
cow_n_col |
cowplot param: how many columns |
cow_rel_h |
cowplot param: relative height |
cow_rel_w |
cowplot param: relative width |
cow_align |
cowplot param: how to align |
show_legend |
show legend |
legend_text |
size of legend text |
legend_symbol_size |
size of legend symbols |
dim_background_color |
background color of points in dim. reduction space |
spat_background_color |
background color of spatial points |
vor_border_color |
border color for voronoi plot |
vor_max_radius |
maximum radius for voronoi ‘cells’ |
vor_alpha |
transparency of voronoi ‘cells’ |
axis_text |
size of axis text |
axis_title |
size of axis title |
show_plot |
show plot |
return_plot |
return ggplot object |
save_plot |
directly save the plot [boolean] |
save_param |
list of saving parameters, see showSaveParameters() |
default_save_name |
default save name for saving, don’t change, change save_name in save_param |
Value¶
A ggplot.
Details¶
Description of parameters …
See also
For other spatial and dimension reduction cell annotation visualizations see: spatDimCellPlot2D()
Examples¶
data(mini_giotto_single_cell)
# combine all metadata
combineMetadata(mini_giotto_single_cell, spat_enr_names = 'cluster_metagene')
#> cell_ID nr_genes perc_genes total_expr leiden_clus cell_types sdimx
#> 1: cell_2 13 65 111.98320 3 cell C 1589.47
#> 2: cell_7 15 75 115.73030 3 cell C 1291.34
#> 3: cell_12 11 55 95.49802 1 cell A 1183.07
#> 4: cell_15 12 60 99.94782 3 cell C 1115.86
#> 5: cell_17 13 65 111.32963 2 cell B 1074.92
#> 6: cell_30 11 55 96.64302 3 cell C 882.00
#> 7: cell_37 6 30 57.77777 2 cell B 618.20
#> 8: cell_40 9 45 82.84693 2 cell B 565.40
#> 9: cell_44 9 45 79.93838 2 cell B 417.40
#> 10: cell_53 9 45 82.40747 1 cell A 1831.19
#> 11: cell_64 8 40 73.06345 1 cell A 1839.07
#> 12: cell_74 11 55 93.04295 3 cell C 1575.84
#> 13: cell_85 8 40 73.72574 1 cell A 1440.75
#> 14: cell_86 14 70 115.75186 1 cell A 1427.06
#> 15: cell_90 11 55 93.02181 1 cell A 1351.50
#> 16: cell_95 6 30 59.55714 1 cell A 1228.13
#> 17: cell_96 10 50 88.31757 1 cell A 1210.65
#> 18: cell_107 16 80 130.62640 1 cell A 969.60
#> 19: cell_113 12 60 99.83100 2 cell B 874.30
#> 20: cell_118 14 70 117.63523 2 cell B 270.00
#> sdimy 1 2 3
#> 1: -669.51 3.144429 8.617638 5.853656
#> 2: -957.71 4.088076 9.410168 4.427447
#> 3: -950.97 2.899783 9.264667 2.785292
#> 4: -1021.40 4.058155 7.842009 3.405087
#> 5: -391.16 6.413588 7.374390 2.629099
#> 6: -668.36 2.989329 9.298030 2.823368
#> 7: -894.70 7.222222 0.000000 0.000000
#> 8: -421.27 5.933558 3.031319 2.865092
#> 9: -669.71 8.067155 0.000000 2.566856
#> 10: -1090.20 2.183105 6.374428 4.449344
#> 11: -1458.00 0.985555 9.382938 1.480231
#> 12: -1829.60 1.715689 8.215992 5.003582
#> 13: -1298.30 0.000000 7.914246 4.373377
#> 14: -1401.00 3.790383 5.580052 8.658080
#> 15: -1923.80 1.839913 9.190628 3.859789
#> 16: -739.38 2.523159 6.561978 0.000000
#> 17: -374.81 3.737206 8.241875 1.494778
#> 18: -1198.50 4.579634 8.674903 6.989984
#> 19: -1127.00 5.564253 7.927811 1.291685
#> 20: -1383.30 9.142231 1.263504 6.152727
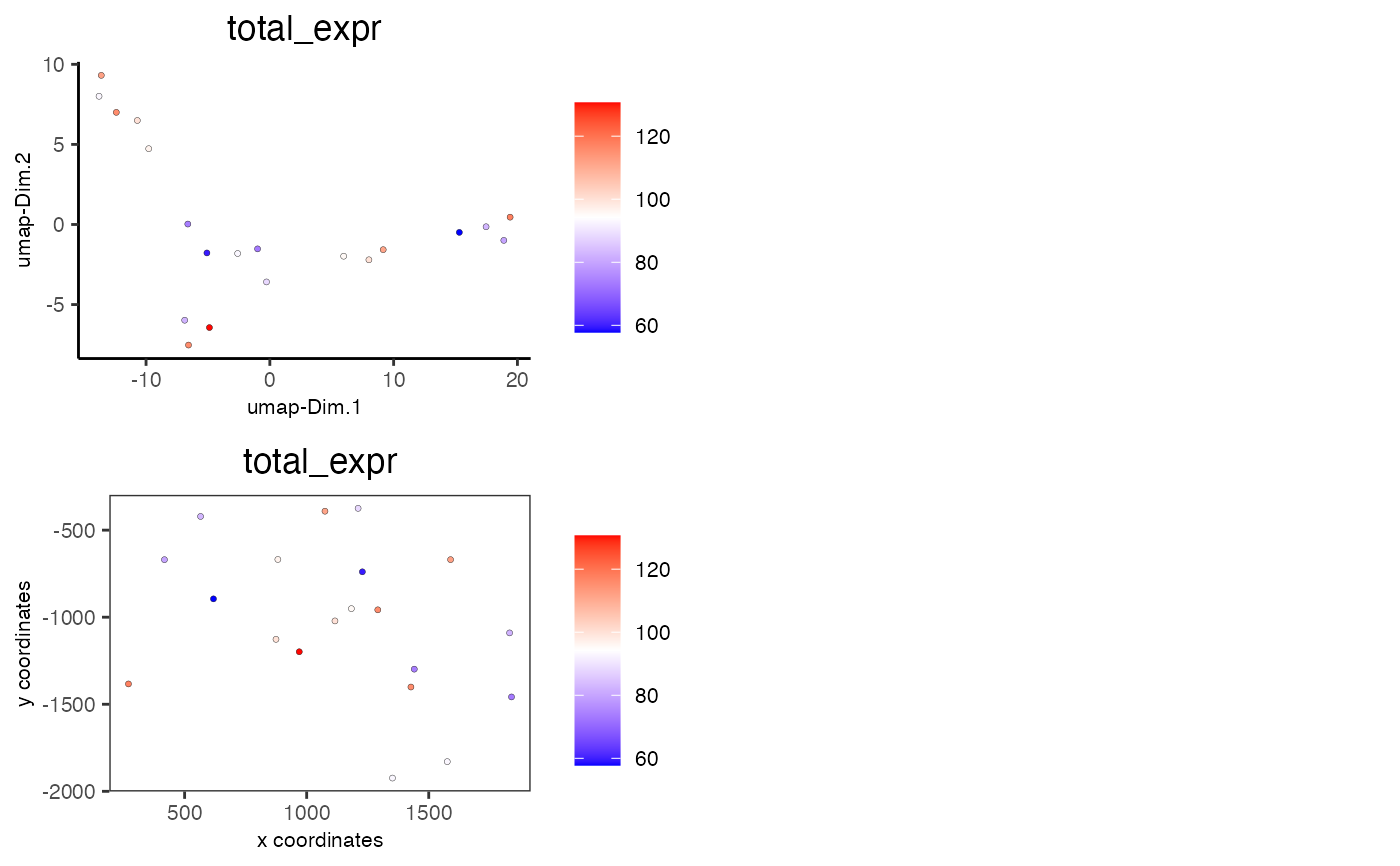
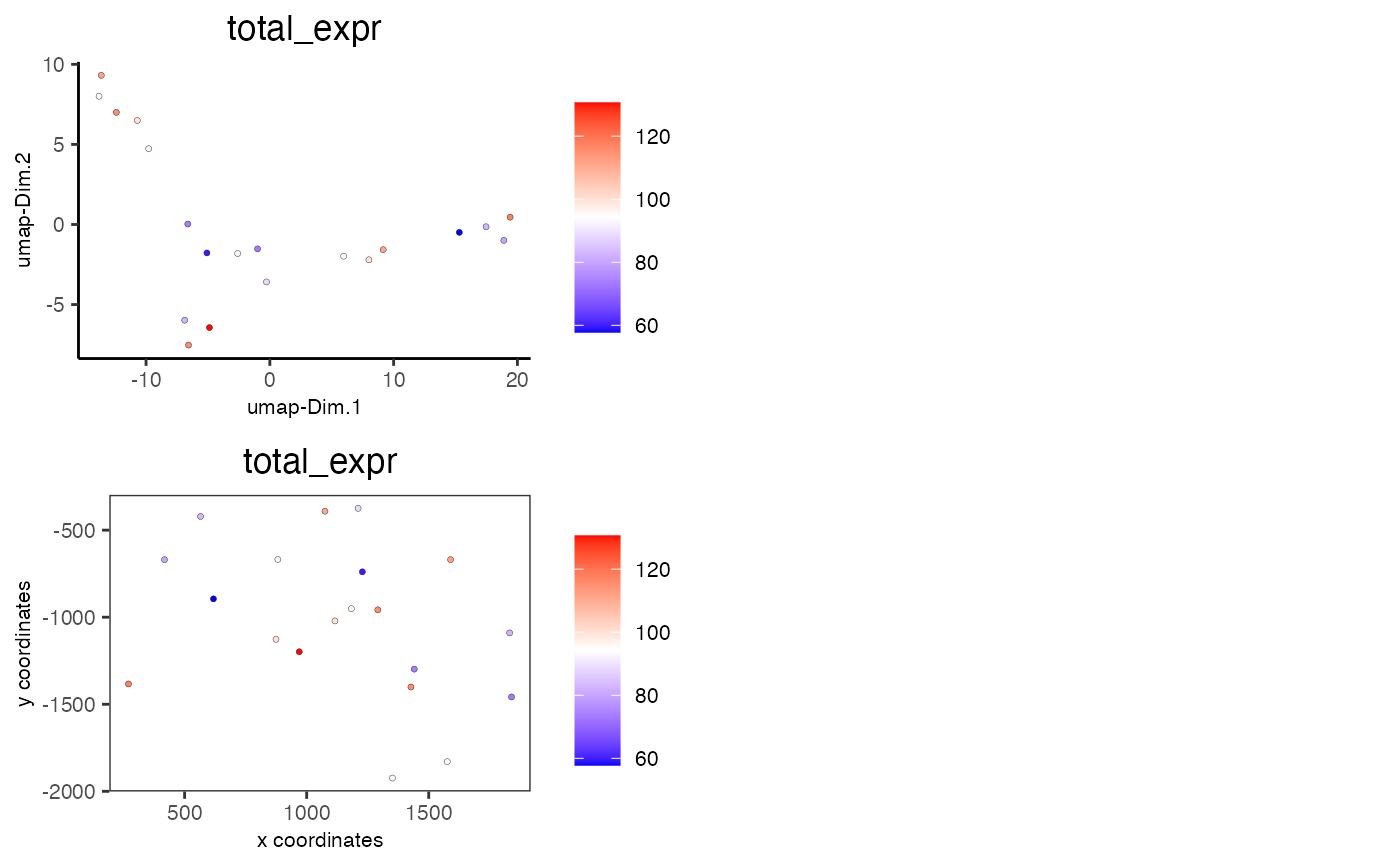
# visualize total expression information
spatDimCellPlot(mini_giotto_single_cell, cell_annotation_values = 'total_expr')
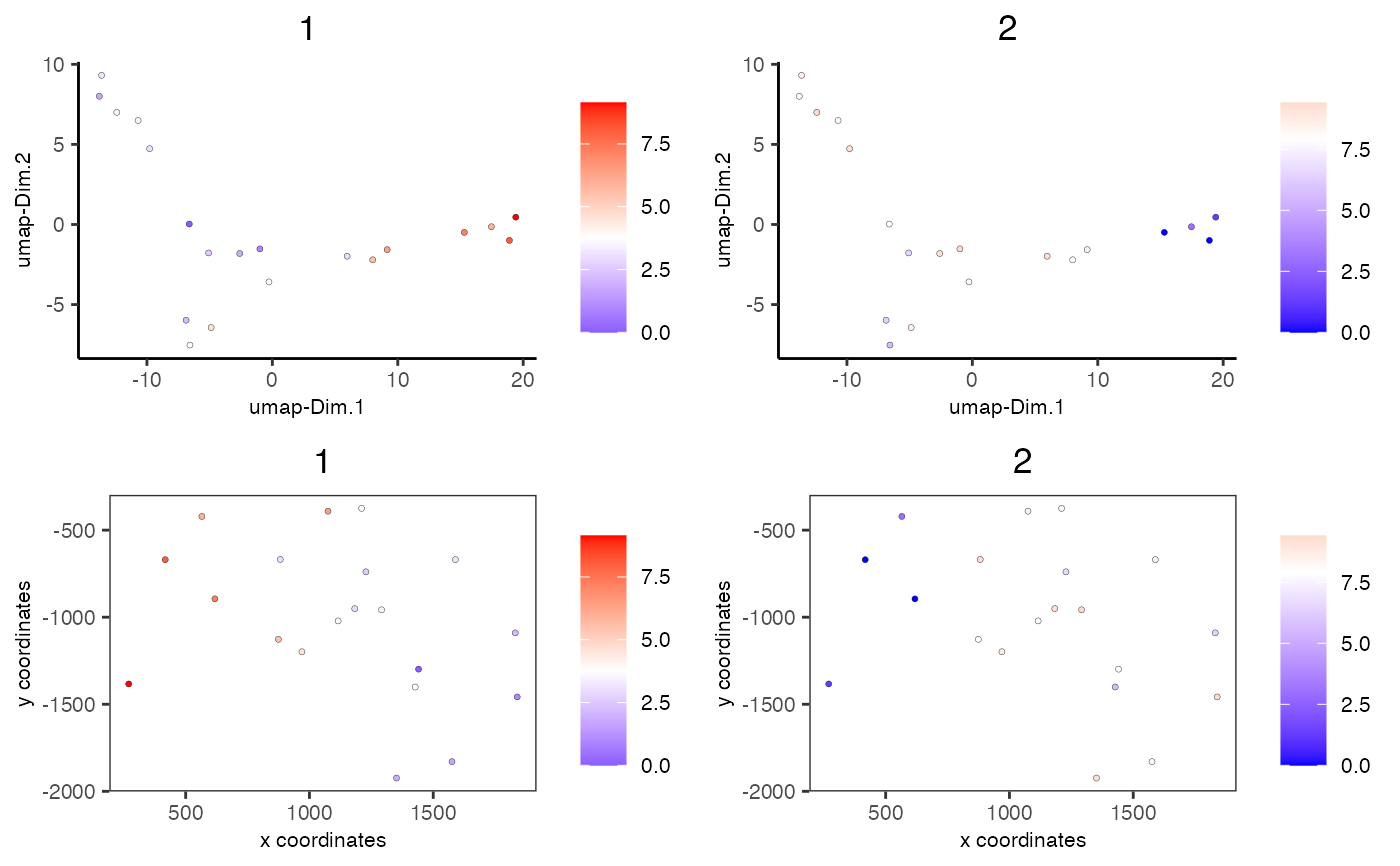
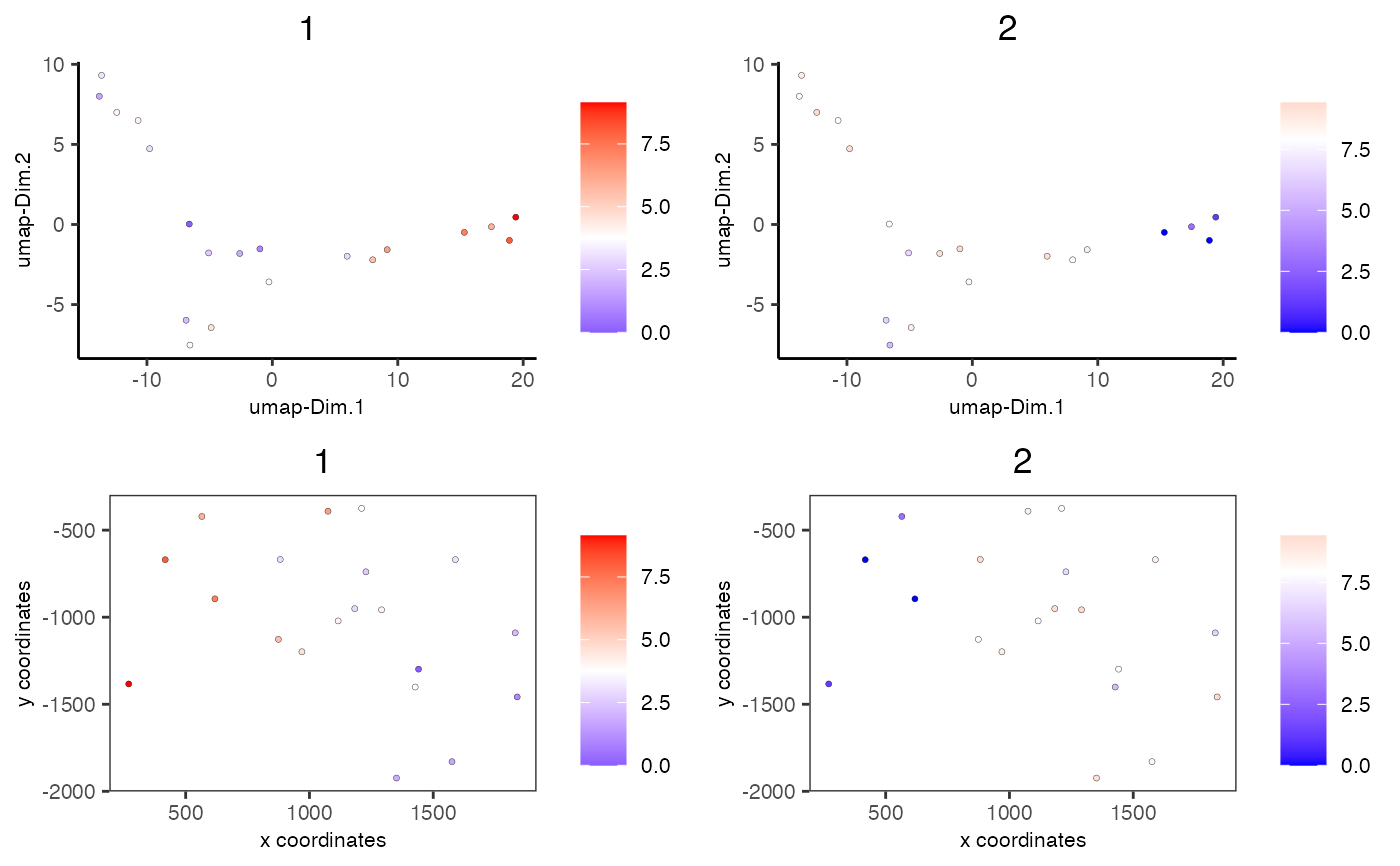
# visualize enrichment results
spatDimCellPlot(mini_giotto_single_cell,
spat_enr_names = 'cluster_metagene',
cell_annotation_values = c('1','2'))