Mouse osmFISH SS Cortex¶
Warning
This tutorial was written with Giotto version 0.3.6.9046, your version is 1.0.3. This is a more recent version and results should be reproducible.
Install Python and R Modules¶
To run this vignette you need to install all of the necessary Python modules.
Important
Python module installation can be done either automatically via our installation tool (from within R) (see step 2.2A) or manually (see step 2.2B).
See Part 2.2 Giotto-Specific Python Packages of our Giotto Installation section for step-by-step instructions.
Set-Up Giotto¶
library(Giotto)
Set A Working Directory¶
#results_folder = '/path/to/directory/'
results_folder = '/Volumes/Ruben_Seagate/Dropbox (Personal)/Projects/GC_lab/Ruben_Dries/190225_spatial_package/Results/Visium/Brain/201226_results//'
Set A Giotto Python Path¶
# set python path to your preferred python version path
# set python path to NULL if you want to automatically install (only the 1st time) and use the giotto miniconda environment
python_path = NULL
if(is.null(python_path)) {
installGiottoEnvironment()
}
Dataset Explanation¶
Codeluppi et al. created a cyclic single-molecule fluorescence in situ hybridization (osmFISH) technology and define the cellular organization of the somatosensory cortex with the expression of 33 genes in 5,328 cells.
1. Giotto Global Instructions and Preparations¶
## instructions allow us to automatically save all plots into a chosen results folder
instrs = createGiottoInstructions(save_plot = TRUE,
show_plot = FALSE,
save_dir = results_folder,
python_path = python_path)
expr_path = paste0(results_folder, "osmFISH_prep_expression.txt")
loc_path = paste0(results_folder, "osmFISH_prep_cell_coordinates.txt")
meta_path = paste0(results_folder, "osmFISH_prep_cell_metadata.txt")
2. Create Giotto Object and Process The Data¶
## create
osm_test <- createGiottoObject(raw_exprs = expr_path,
spatial_locs = loc_path,
instructions = instrs)
showGiottoInstructions(osm_test)
## add field annotation
metadata = data.table::fread(file = meta_path)
osm_test = addCellMetadata(osm_test, new_metadata = metadata,
by_column = T, column_cell_ID = 'CellID')
## filter
osm_test <- filterGiotto(gobject = osm_test,
expression_threshold = 1,
gene_det_in_min_cells = 10,
min_det_genes_per_cell = 10,
expression_values = c('raw'),
verbose = T)
## normalize
# 1. standard z-score way
osm_test <- normalizeGiotto(gobject = osm_test)
# 2. osmFISH way
raw_expr_matrix = osm_test@raw_exprs
norm_genes = (raw_expr_matrix/rowSums_giotto(raw_expr_matrix)) * nrow(raw_expr_matrix)
norm_genes_cells = t_giotto((t_giotto(norm_genes)/colSums_giotto(norm_genes)) * ncol(raw_expr_matrix))
osm_test@custom_expr = norm_genes_cells
## add gene & cell statistics
osm_test <- addStatistics(gobject = osm_test)
## add gene & cell statistics
osm_test <- addStatistics(gobject = osm_test)
# save according to giotto instructions
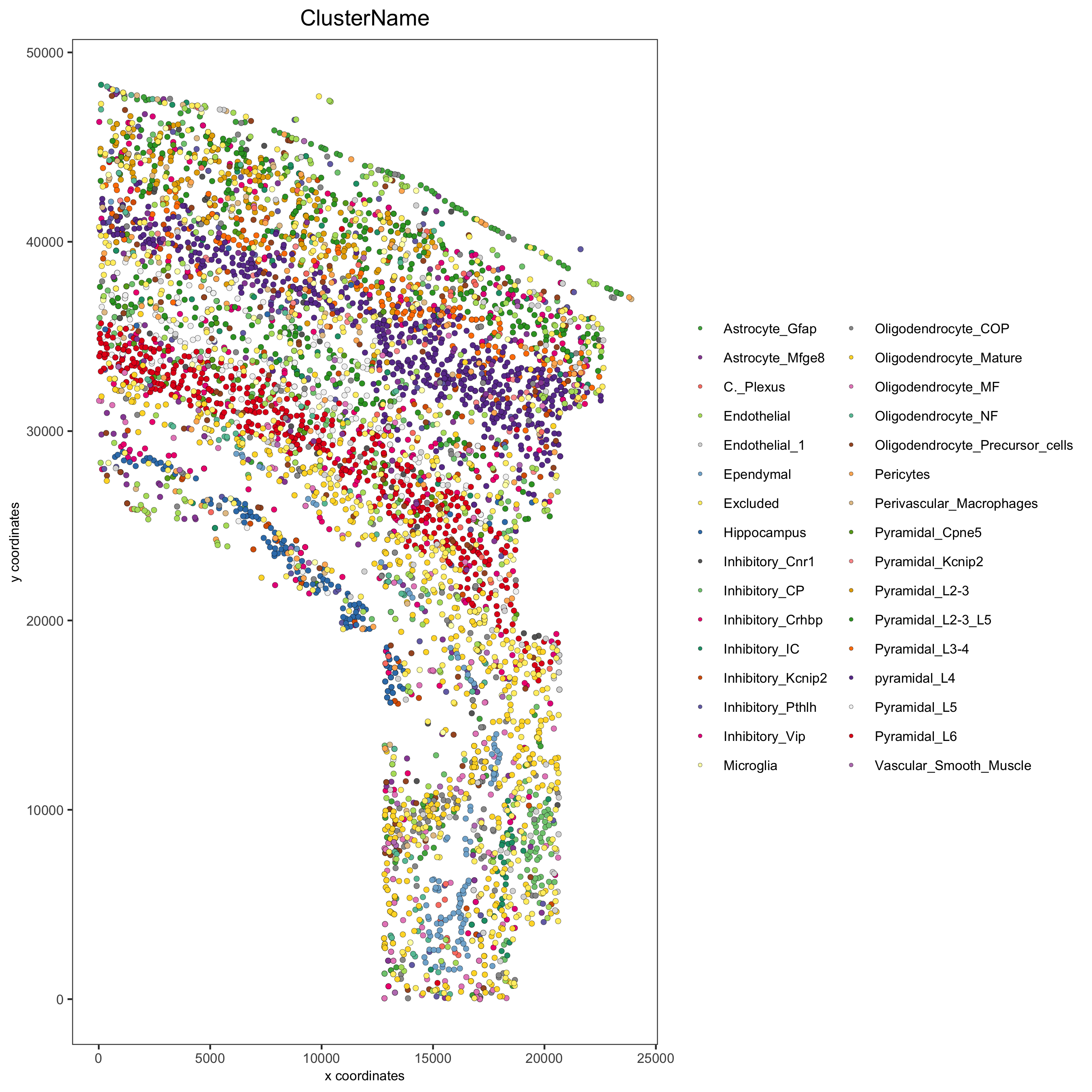
spatPlot(gobject = osm_test, cell_color = 'ClusterName', point_size = 1.5,
save_param = list(save_name = '2_a_original_clusters'))
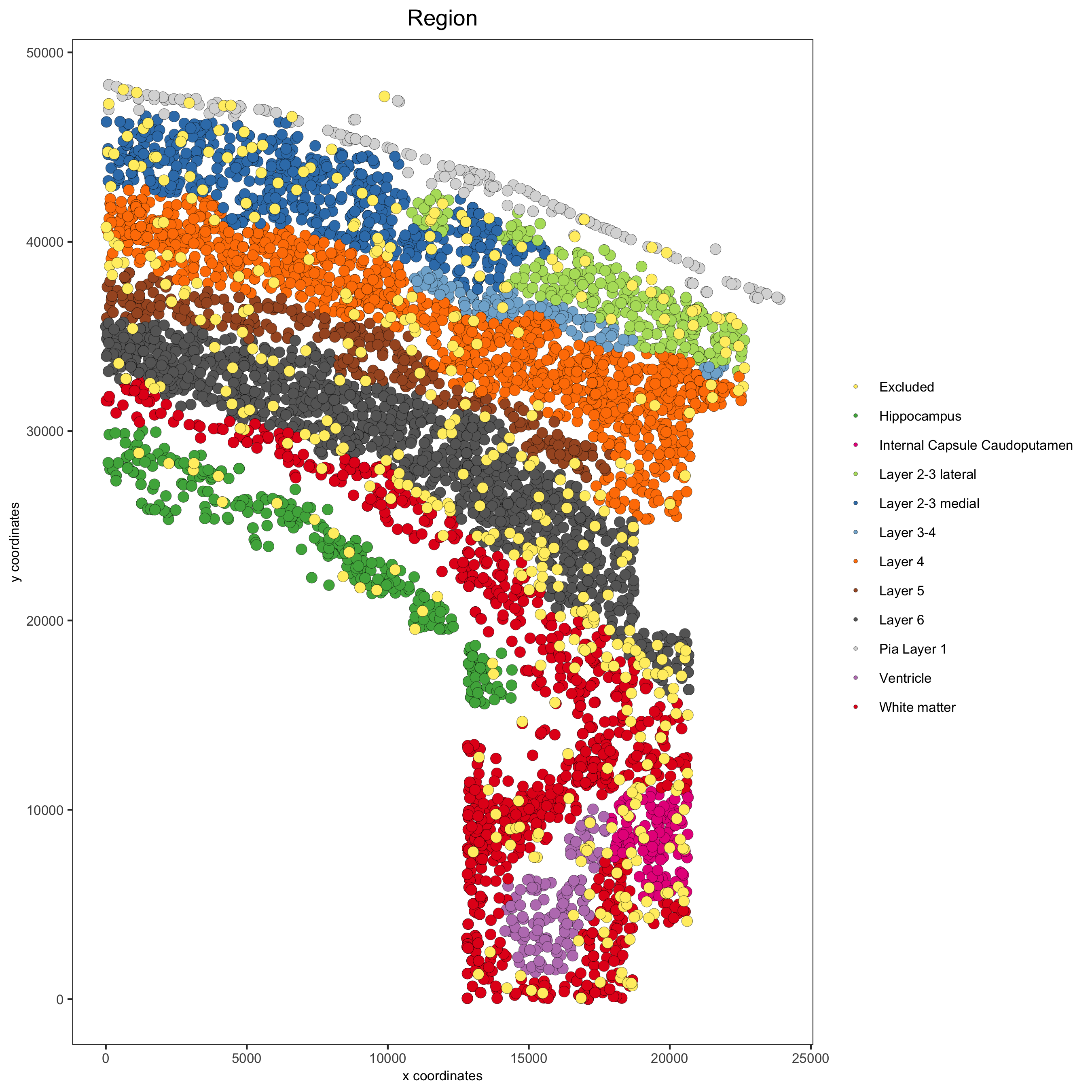
spatPlot(gobject = osm_test, cell_color = 'Region',
save_param = list(save_name = '2_b_original_regions'))
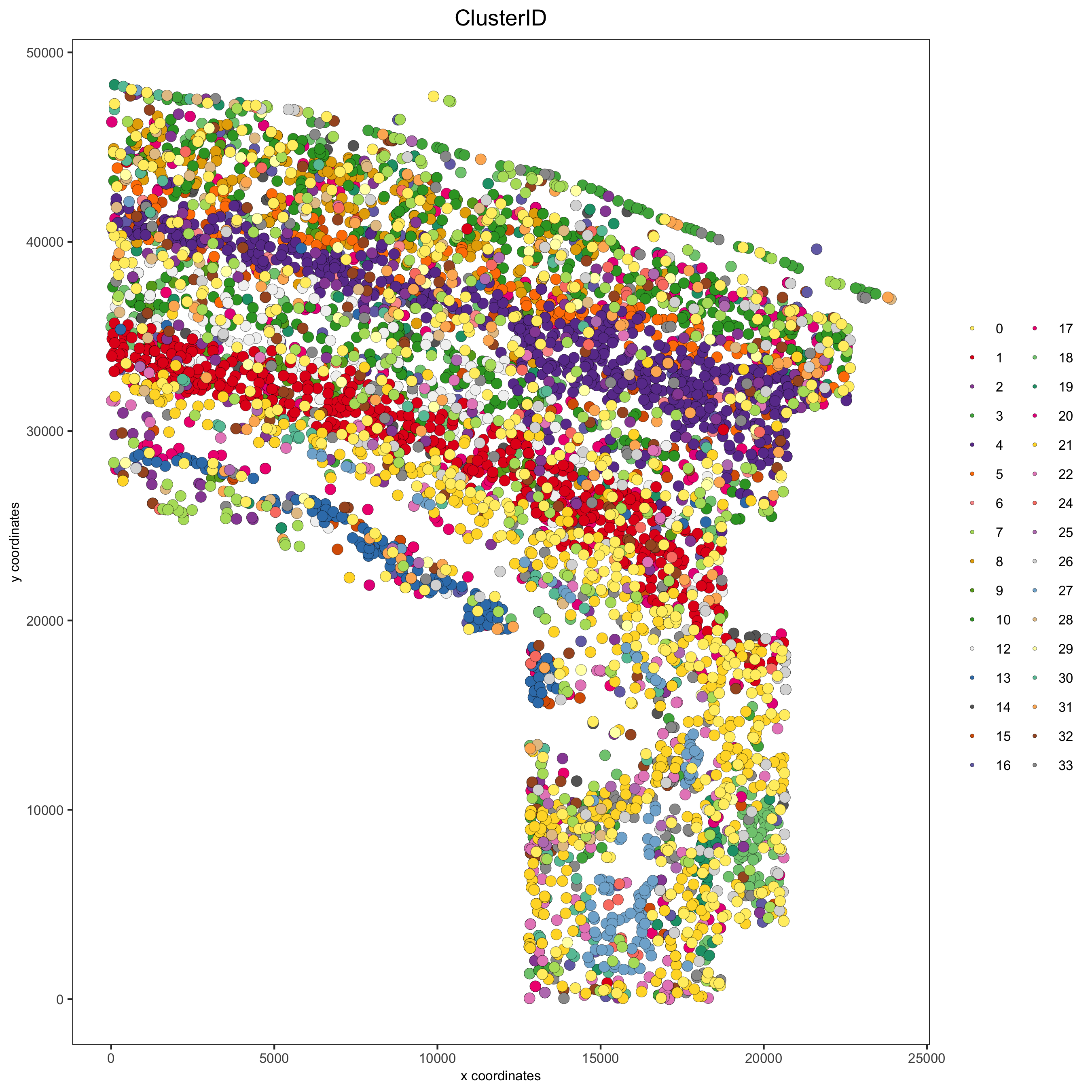
spatPlot(gobject = osm_test, cell_color = 'ClusterID',
save_param = list(save_name = '2_c_clusterID'))
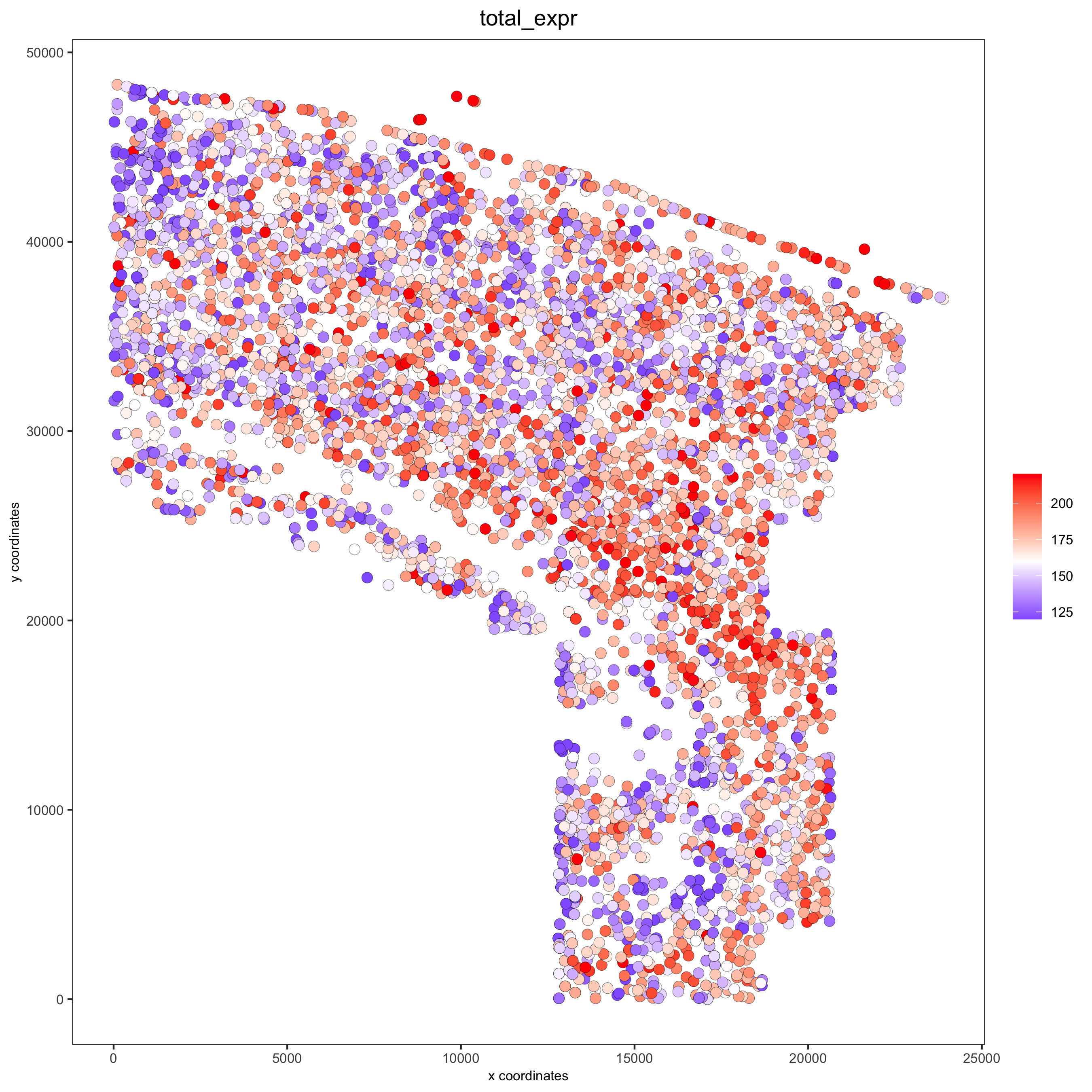
spatPlot(gobject = osm_test, cell_color = 'total_expr', color_as_factor = F, gradient_midpoint = 160,
gradient_limits = c(120,220),
save_param = list(save_name = '2_d_total_expr_limits'))
3. Dimension Reduction¶
## highly variable genes (HVG)
# only 33 genes so use all genes
## run PCA on expression values (default)
osm_test <- runPCA(gobject = osm_test, expression_values = 'custom', scale_unit = F, center = F)
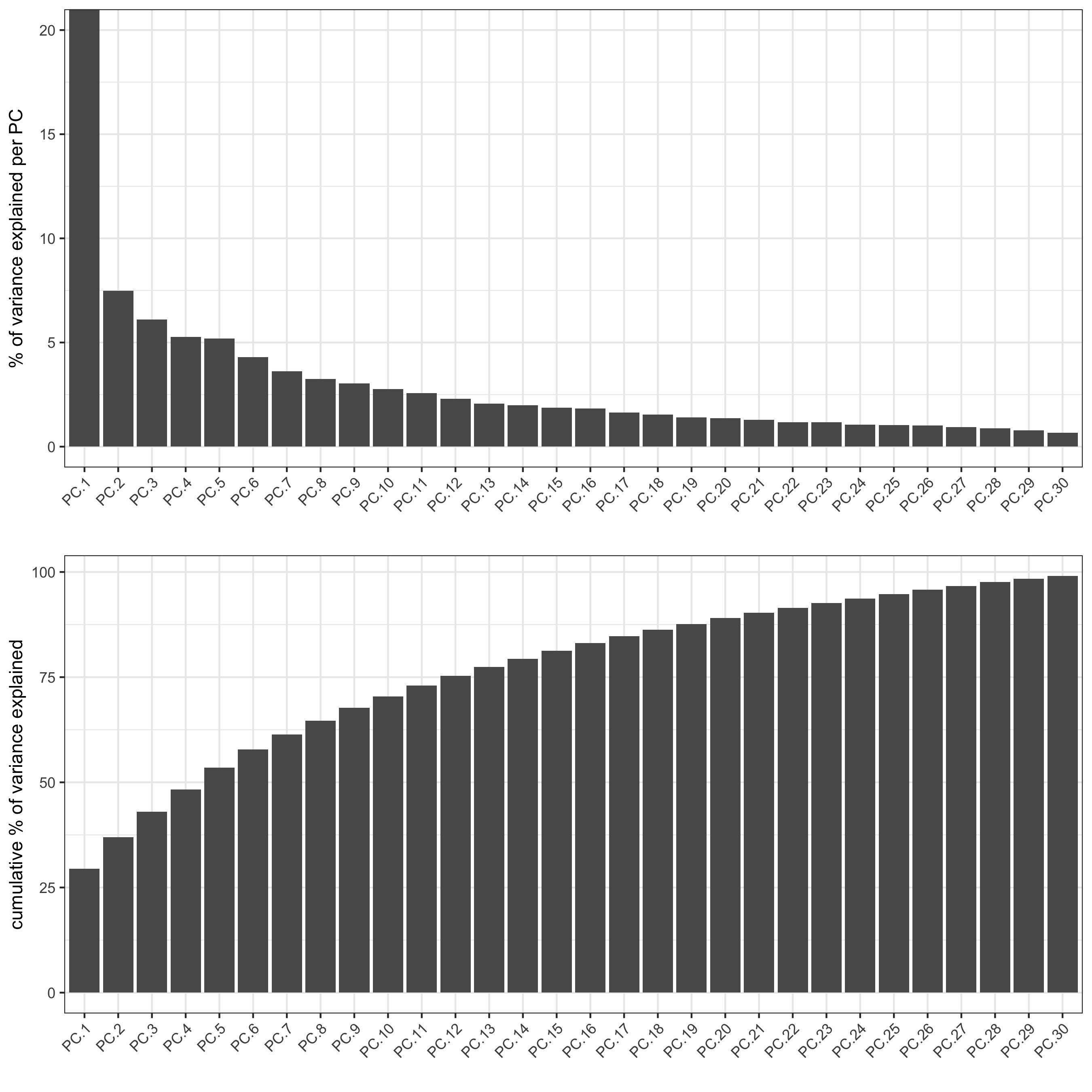
screePlot(osm_test, ncp = 30,
save_param = list(save_name = '3_a_screeplot'))
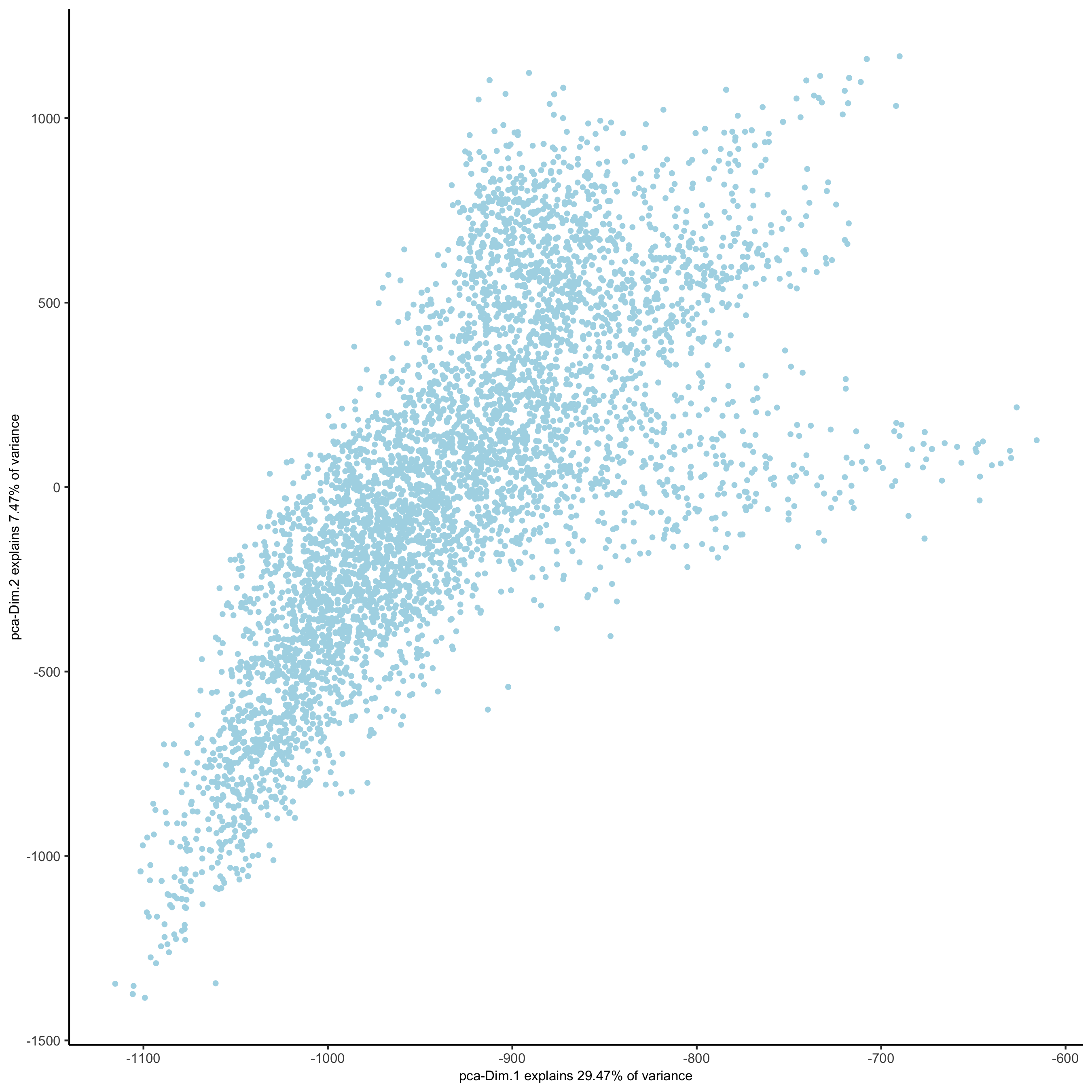
plotPCA(osm_test,
save_param = list(save_name = '3_b_PCA_reduction'))
## run UMAP and tSNE on PCA space (default)
osm_test <- runUMAP(osm_test, dimensions_to_use = 1:31, n_threads = 4)
plotUMAP(gobject = osm_test,
save_param = list(save_name = '3_c_UMAP_reduction.png'))
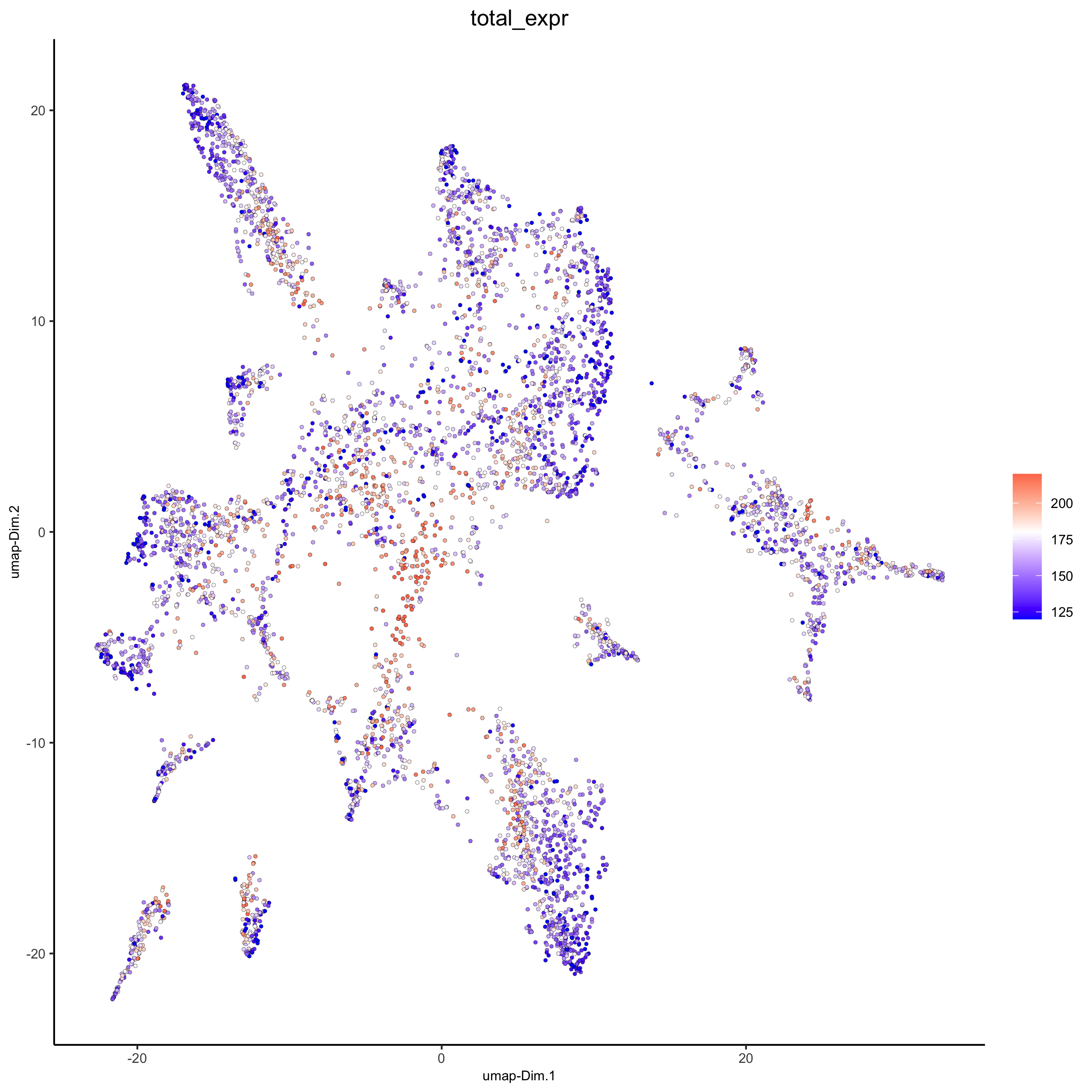
plotUMAP(gobject = osm_test,
cell_color = 'total_expr', color_as_factor = F, gradient_midpoint = 180, gradient_limits = c(120, 220),
save_param = list(save_name = '3_d_UMAP_reduction_expression.png'))
osm_test <- runtSNE(osm_test, dimensions_to_use = 1:31, perplexity = 70, check_duplicates = F)
plotTSNE(gobject = osm_test, save_param = list(save_name = '3_e_tSNE_reduction'))
4. Clustering¶
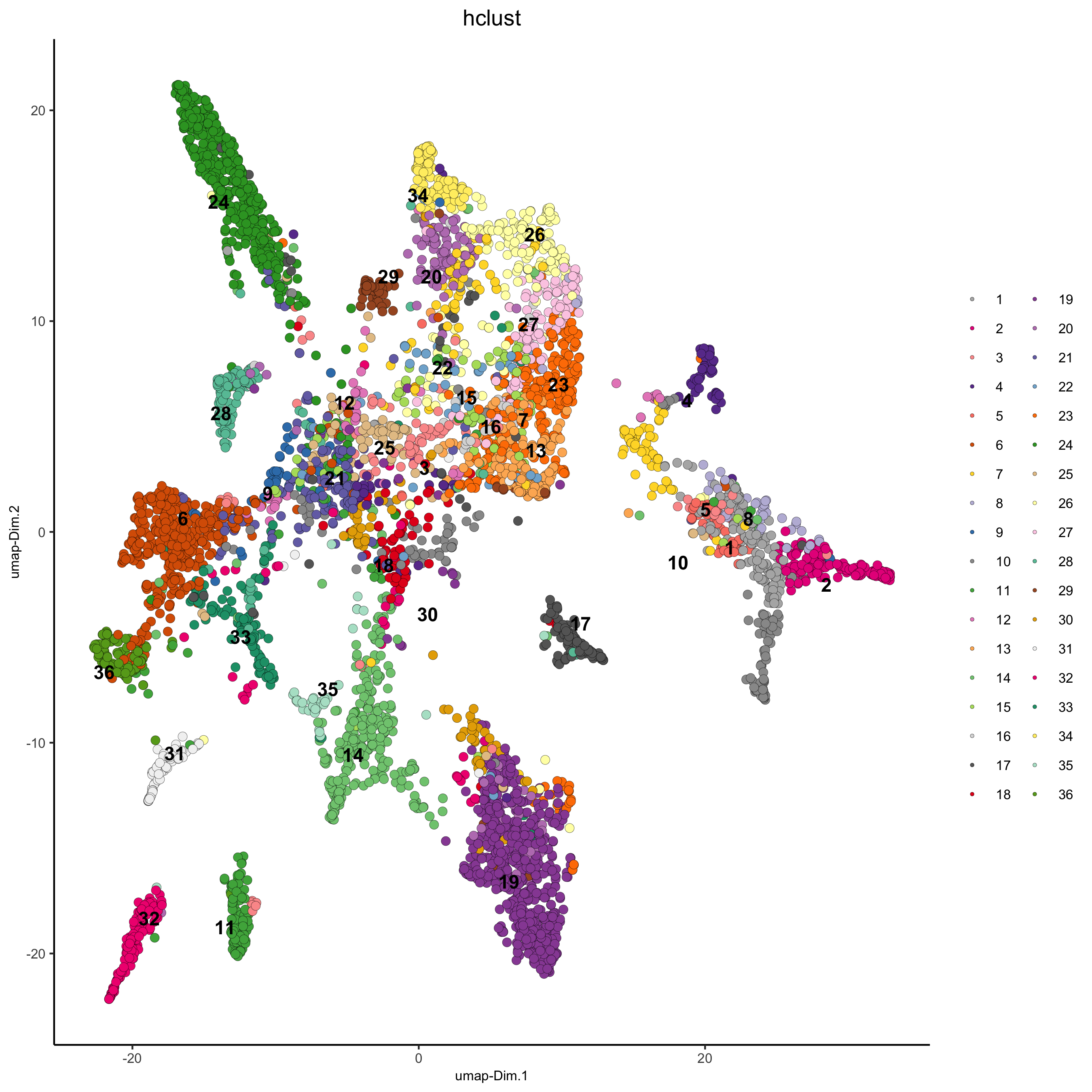
## hierarchical clustering
osm_test = doHclust(gobject = osm_test, expression_values = 'custom', k = 36)
plotUMAP(gobject = osm_test, cell_color = 'hclust', point_size = 2.5,
show_NN_network = F, edge_alpha = 0.05,
save_param = list(save_name = '4_a_UMAP_hclust'))
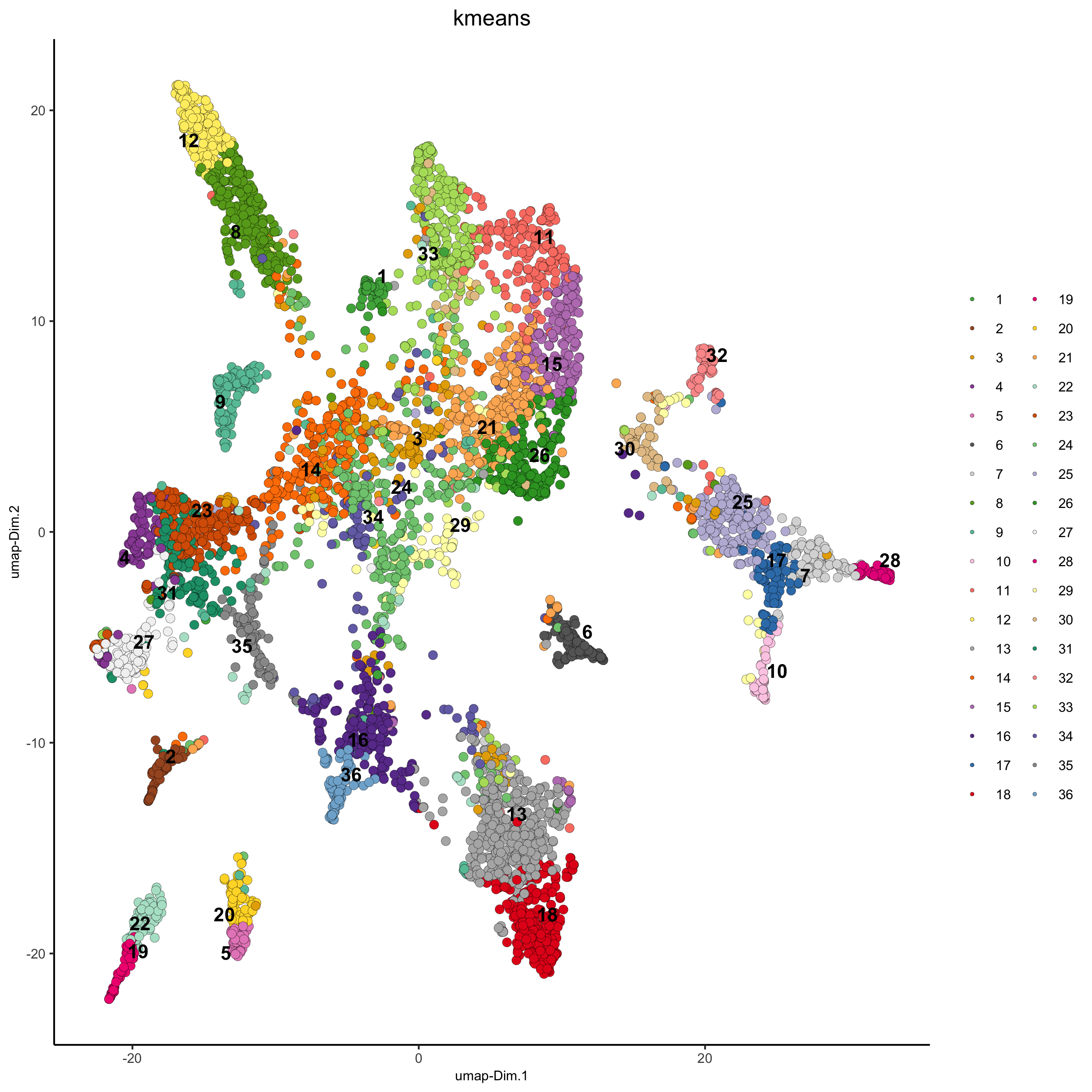
## kmeans clustering
osm_test = doKmeans(gobject = osm_test, dim_reduction_to_use = 'pca', dimensions_to_use = 1:20, centers = 36, nstart = 2000)
plotUMAP(gobject = osm_test, cell_color = 'kmeans',
point_size = 2.5, show_NN_network = F, edge_alpha = 0.05,
save_param = list(save_name = '4_b_UMAP_kmeans'))
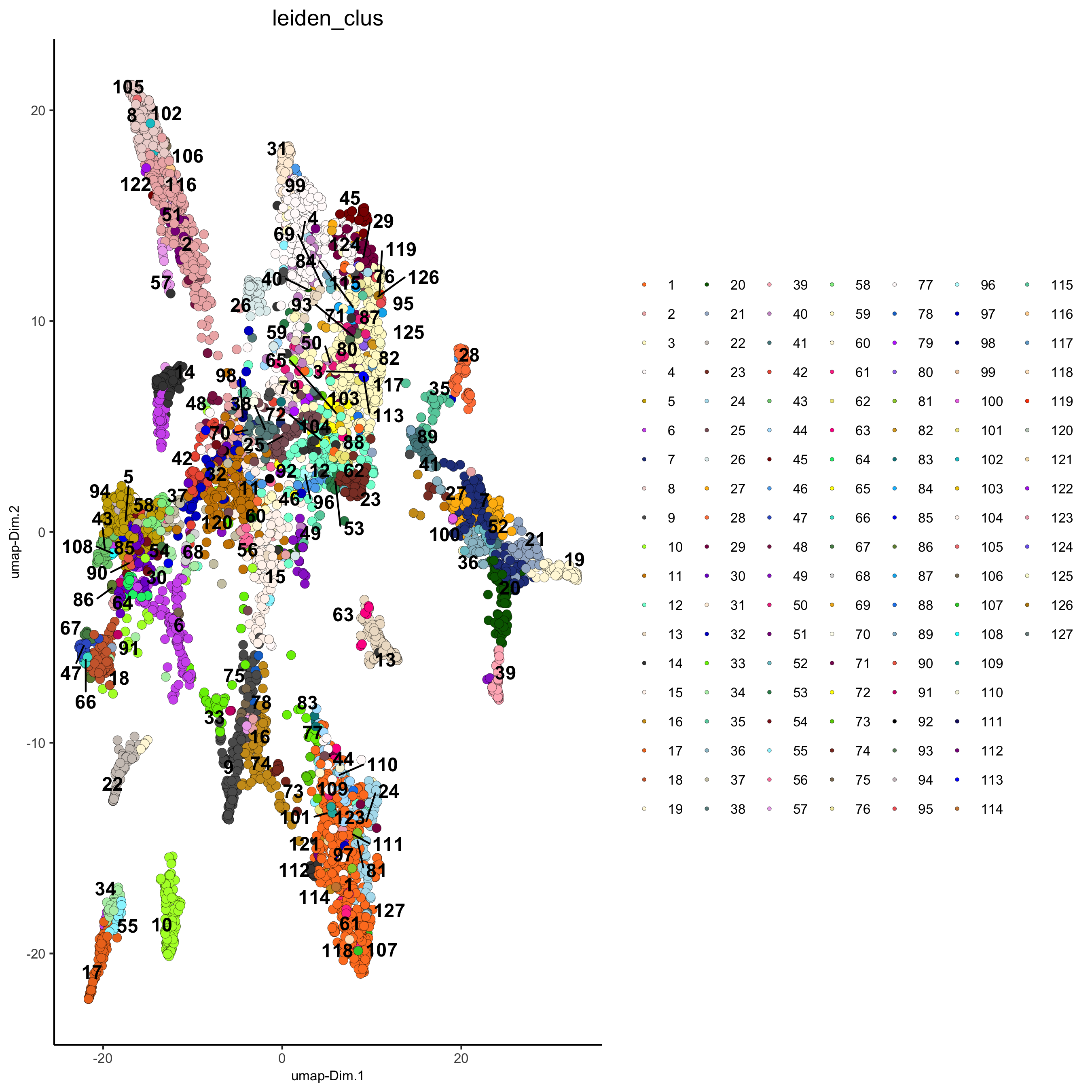
## Leiden clustering strategy:
# 1. overcluster
# 2. merge small clusters that are highly similar
# sNN network (default)
osm_test <- createNearestNetwork(gobject = osm_test, dimensions_to_use = 1:31, k = 12)
osm_test <- doLeidenCluster(gobject = osm_test, resolution = 0.09, n_iterations = 1000)
plotUMAP(gobject = osm_test, cell_color = 'leiden_clus', point_size = 2.5,
show_NN_network = F, edge_alpha = 0.05,
save_param = list(save_name = '4_c_UMAP_leiden'))
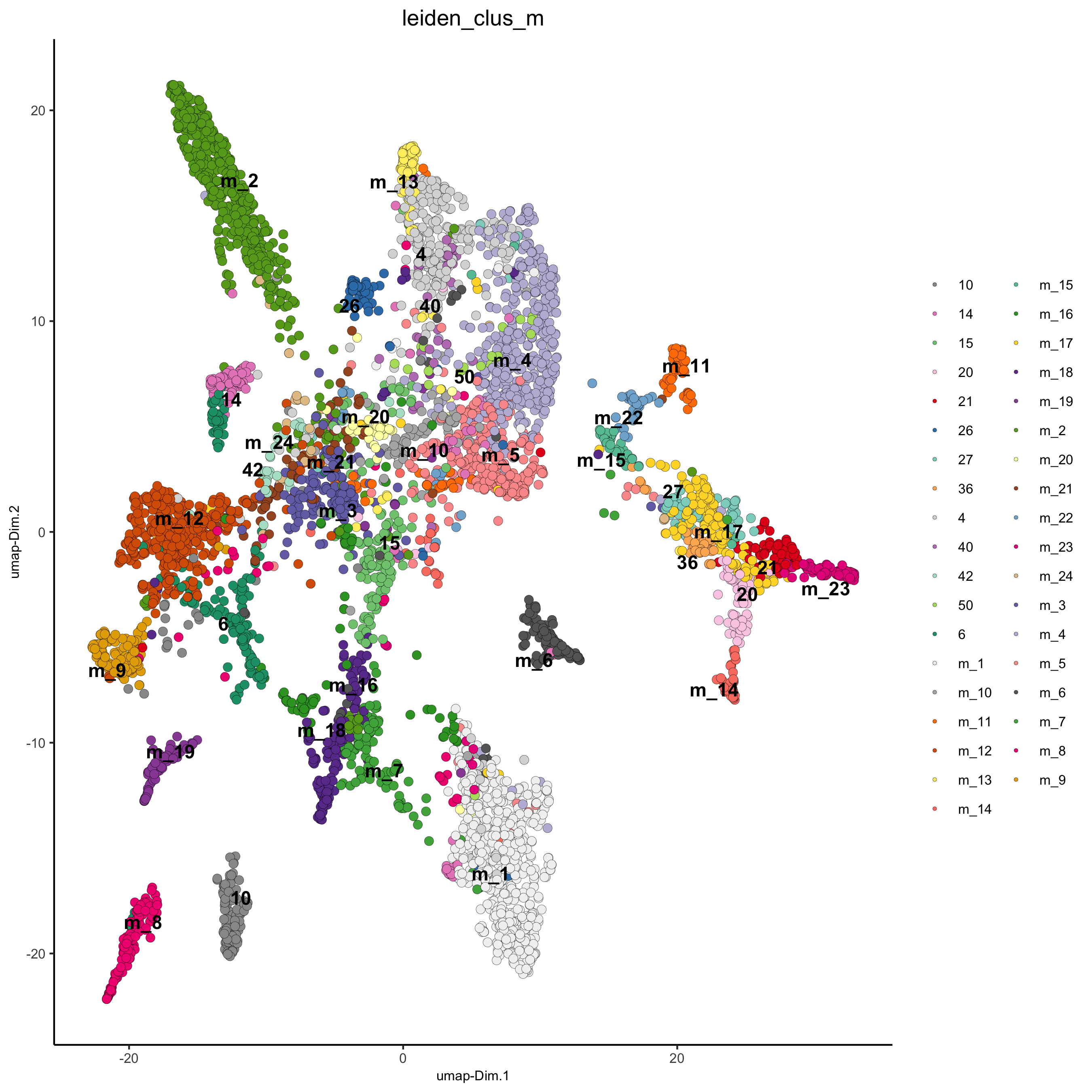
# merge small groups based on similarity
leiden_similarities = getClusterSimilarity(osm_test,
expression_values = 'custom',
cluster_column = 'leiden_clus')
osm_test = mergeClusters(osm_test,
expression_values = 'custom',
cluster_column = 'leiden_clus',
new_cluster_name = 'leiden_clus_m',
max_group_size = 30,
force_min_group_size = 25,
max_sim_clusters = 10,
min_cor_score = 0.7)
plotUMAP(gobject = osm_test, cell_color = 'leiden_clus_m', point_size = 2.5,
show_NN_network = F, edge_alpha = 0.05,
save_param = list(save_name = '4_d_UMAP_leiden_merged'))
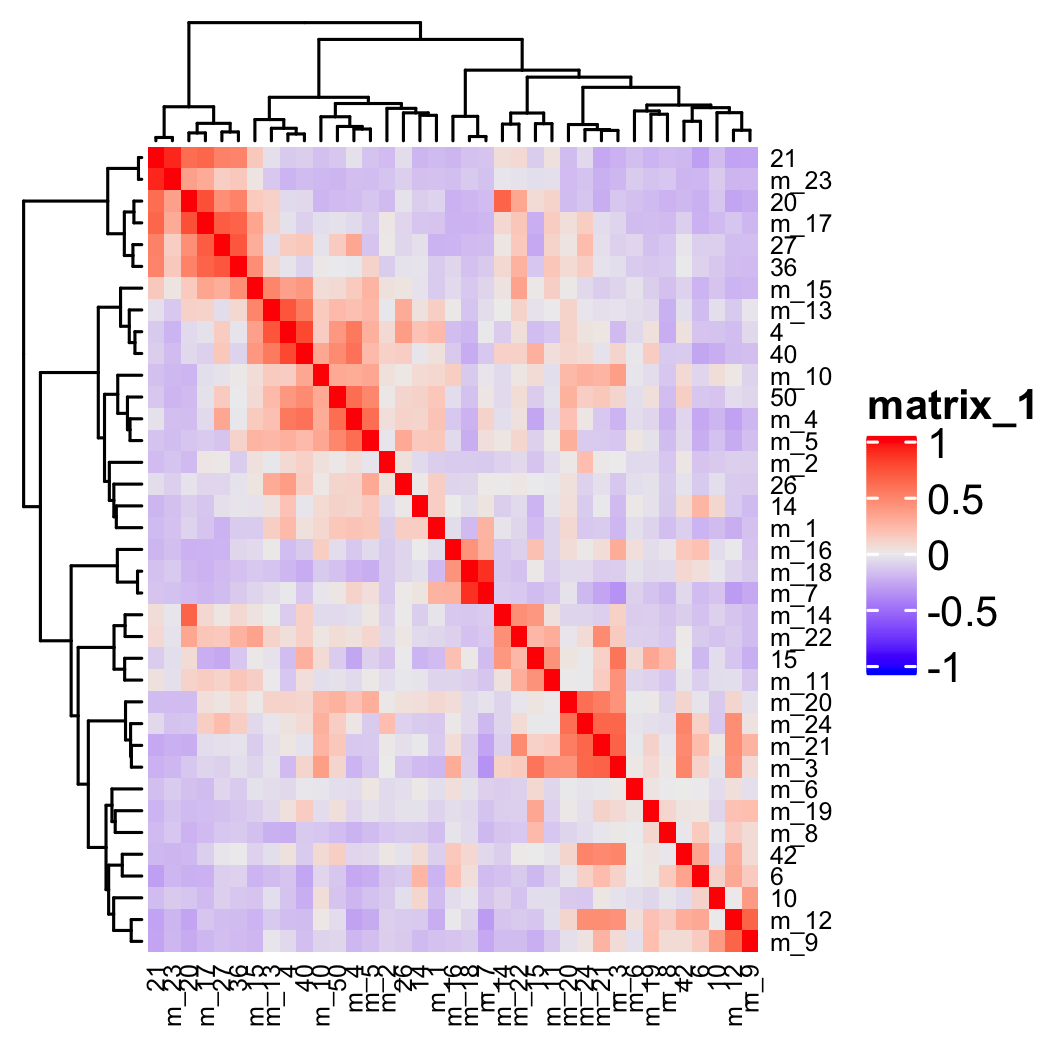
## show cluster relationships
showClusterHeatmap(gobject = osm_test, expression_values = 'custom', cluster_column = 'leiden_clus_m',
save_param = list(save_name = '4_e_heatmap', units = 'cm'),
row_names_gp = grid::gpar(fontsize = 6), column_names_gp = grid::gpar(fontsize = 6))
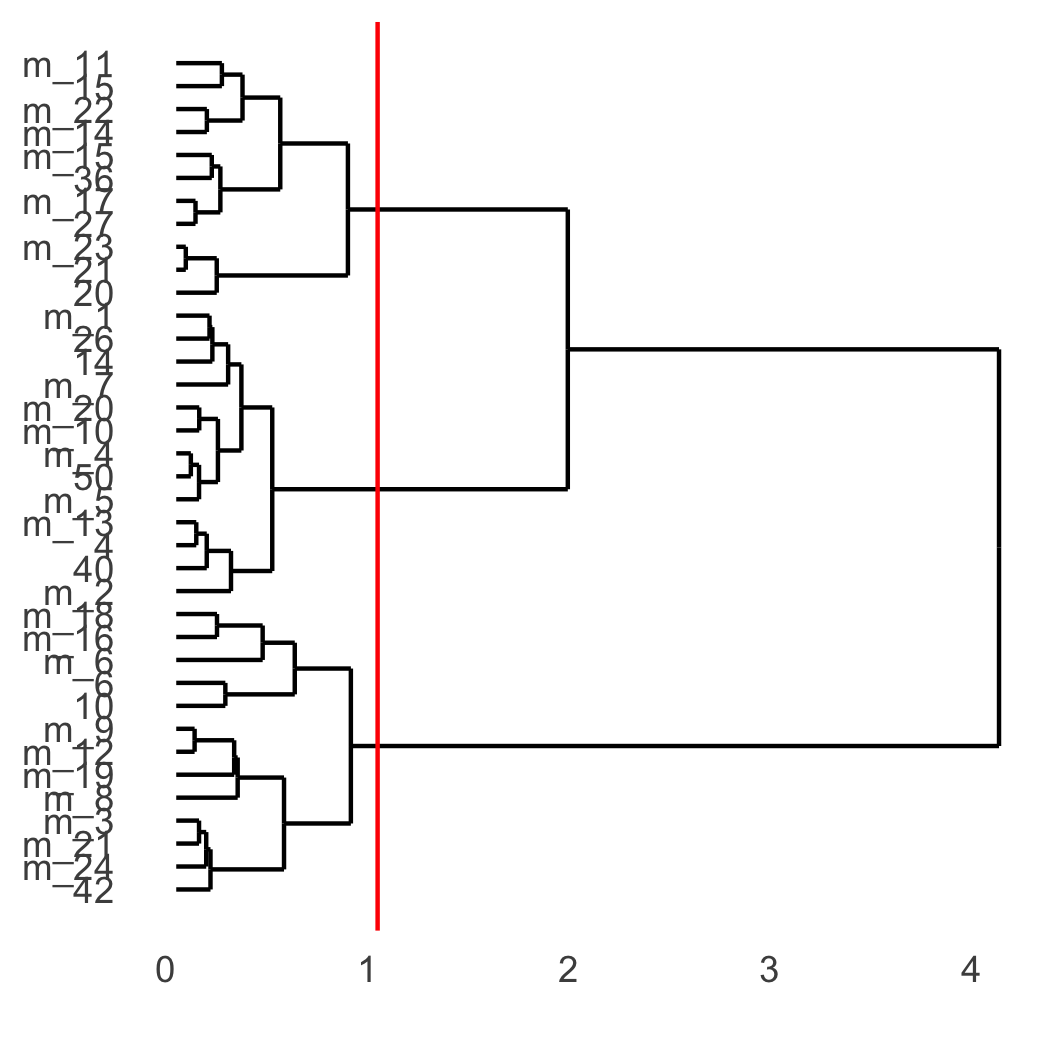
showClusterDendrogram(osm_test, cluster_column = 'leiden_clus_m', h = 1, rotate = T,
save_param = list(save_name = '4_f_dendro', units = 'cm'))
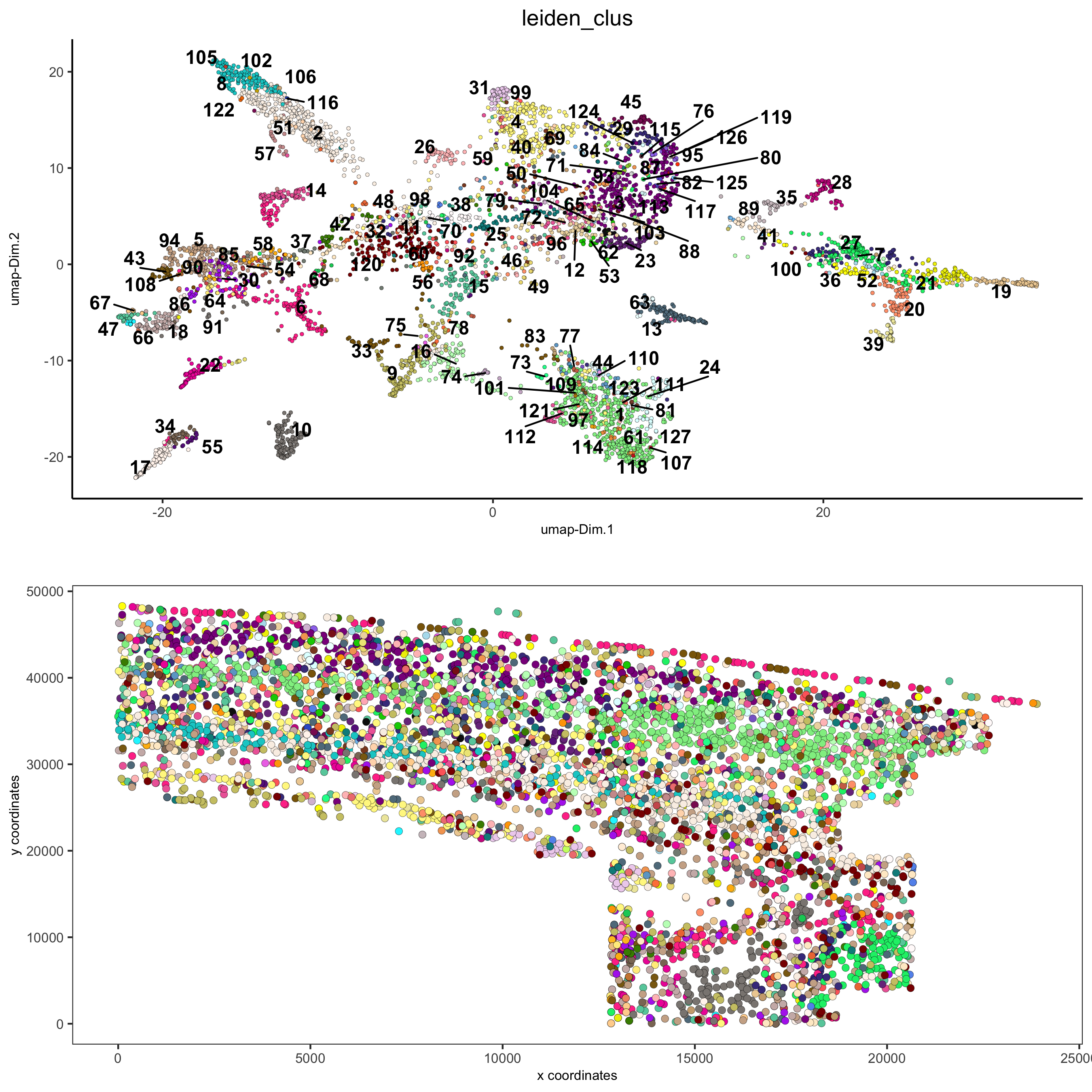
5. Co-Visualization¶
# expression and spatial
spatDimPlot2D(gobject = osm_test, cell_color = 'leiden_clus', spat_point_size = 2,
save_param = list(save_name = '5_a_covis_leiden'))
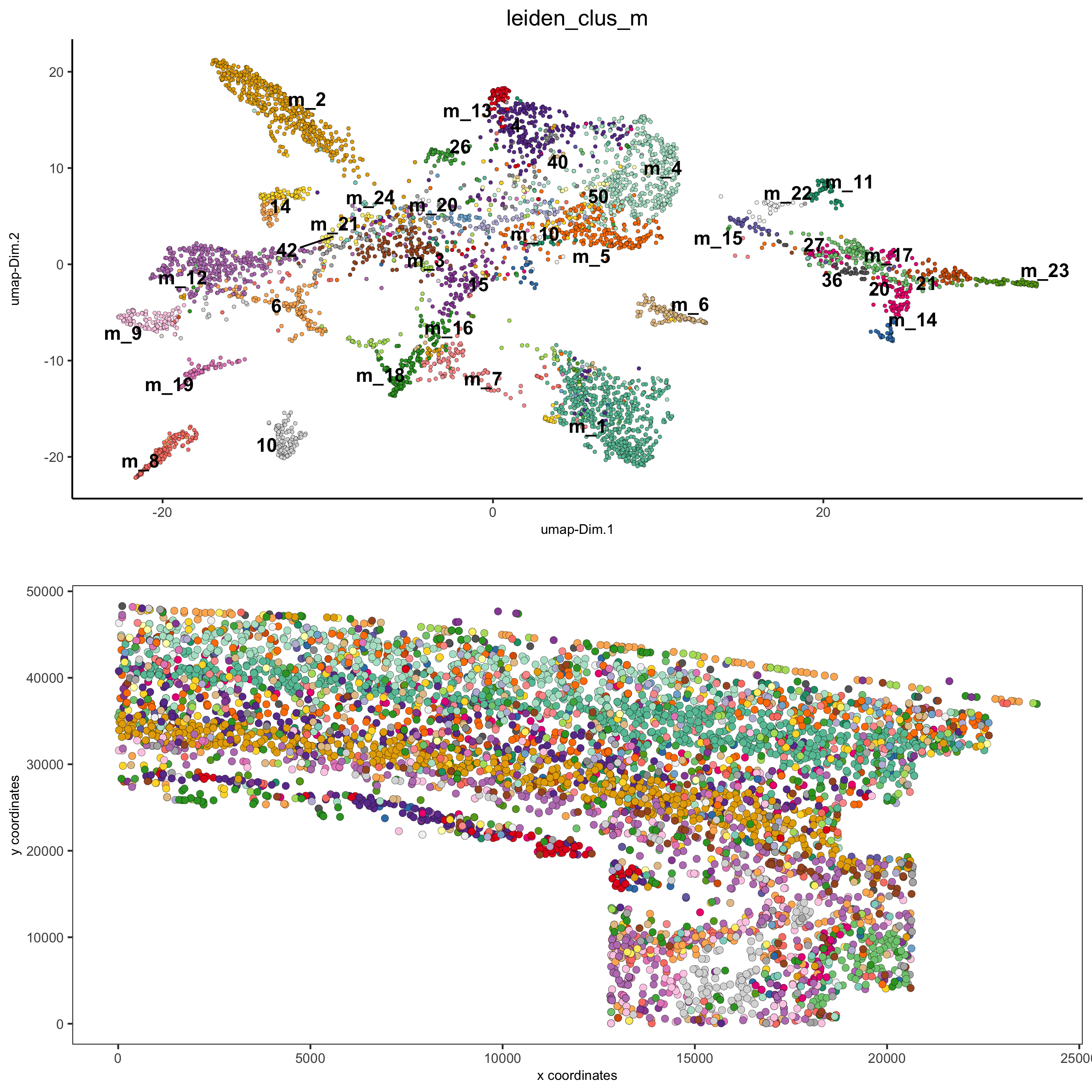
spatDimPlot2D(gobject = osm_test, cell_color = 'leiden_clus_m', spat_point_size = 2,
save_param = list(save_name = '5_b_covis_leiden_m'))
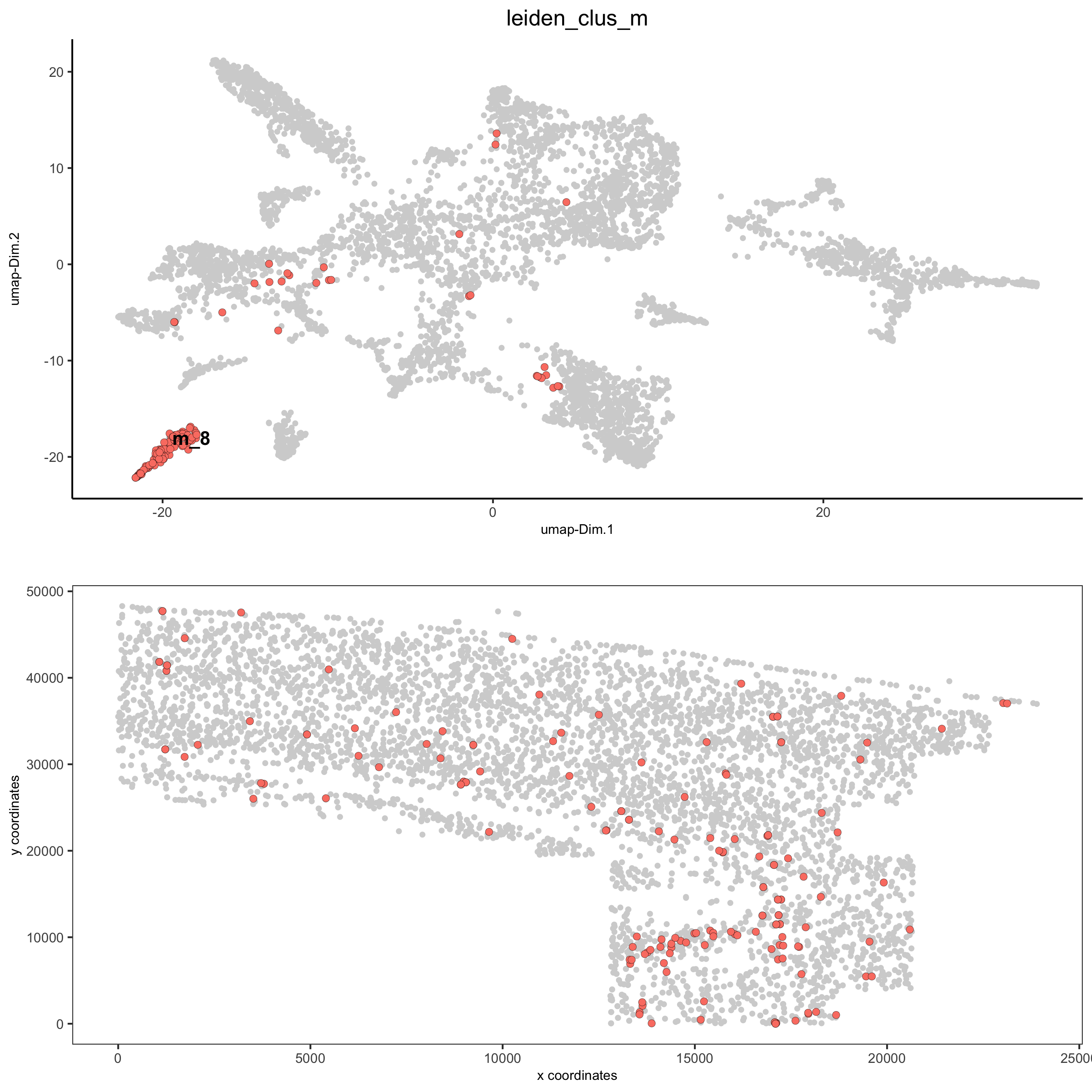
spatDimPlot2D(gobject = osm_test, cell_color = 'leiden_clus_m',
dim_point_size = 2, spat_point_size = 2, select_cell_groups = 'm_8',
save_param = list(save_name = '5_c_covis_leiden_merged_selected'))
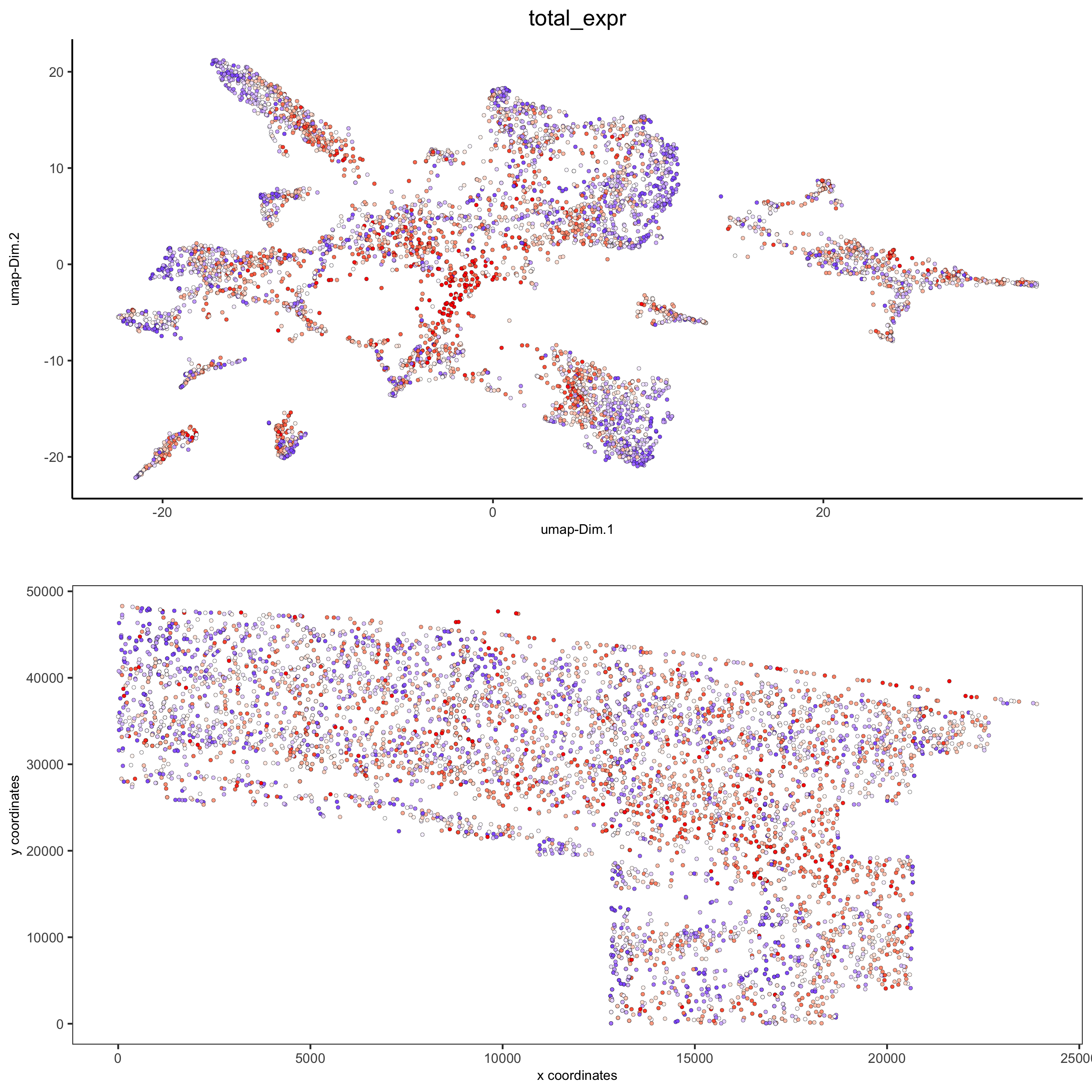
spatDimPlot2D(gobject = osm_test, cell_color = 'total_expr', color_as_factor = F,
gradient_midpoint = 160, gradient_limits = c(120,220),
save_param = list(save_name = '5_d_total_expr'))
6. Differential Expression¶
## split dendrogram nodes ##
dendsplits = getDendrogramSplits(gobject = osm_test,
expression_values = 'custom',
cluster_column = 'leiden_clus_m')
split_3_markers = findGiniMarkers(gobject = osm_test, expression_values = 'custom', cluster_column = 'leiden_clus_m',
group_1 = unlist(dendsplits[3]$tree_1), group_2 = unlist(dendsplits[3]$tree_2))
## Individual populations ##
markers = findMarkers_one_vs_all(gobject = osm_test,
method = 'scran',
expression_values = 'custom',
cluster_column = 'leiden_clus_m',
min_genes = 2, rank_score = 2)
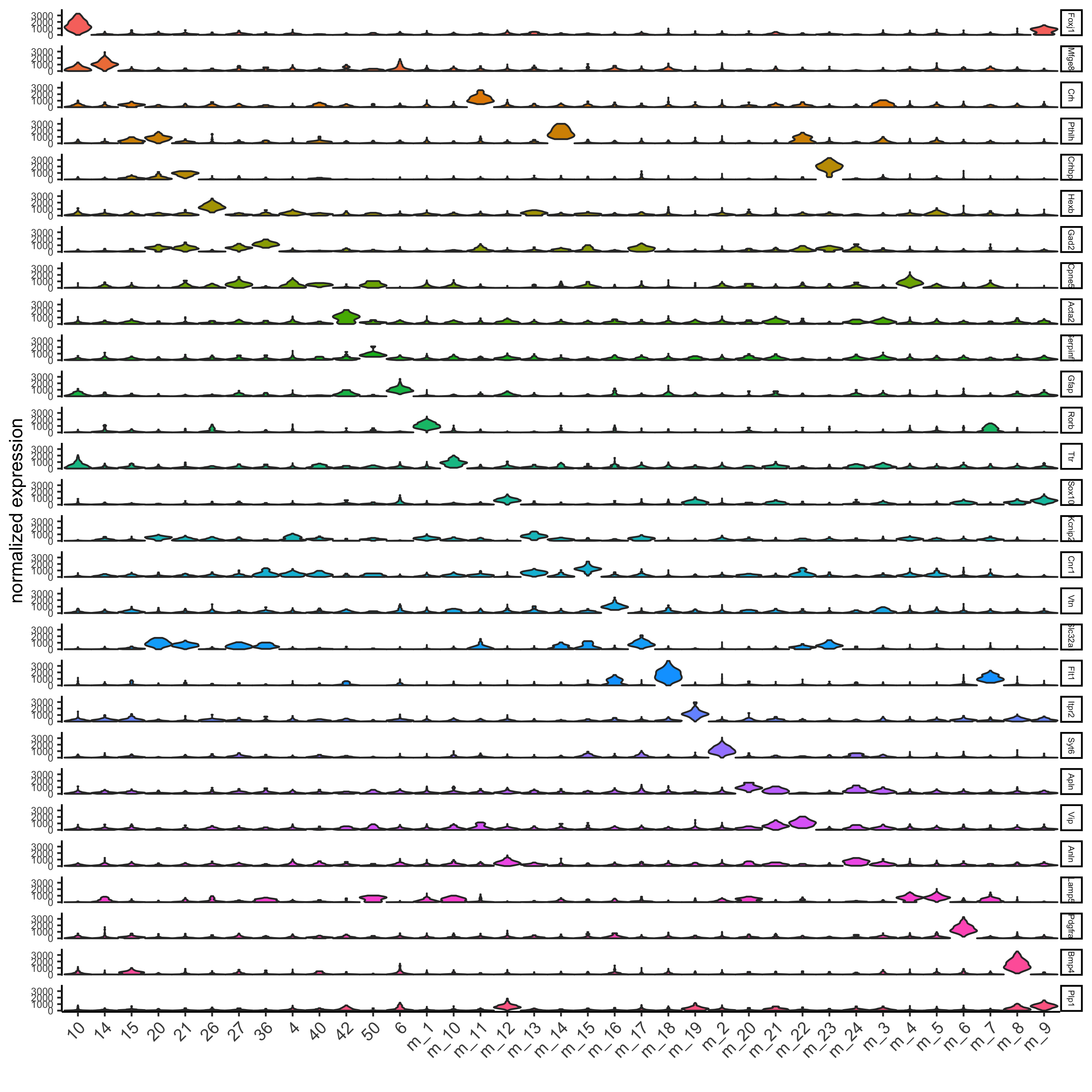
## violinplot
topgenes = markers[, head(.SD, 1), by = 'cluster']$genes
violinPlot(osm_test, genes = unique(topgenes), cluster_column = 'leiden_clus_m', expression_values = 'custom',
strip_text = 5, strip_position = 'right',
save_param = c(save_name = '6_a_violinplot'))
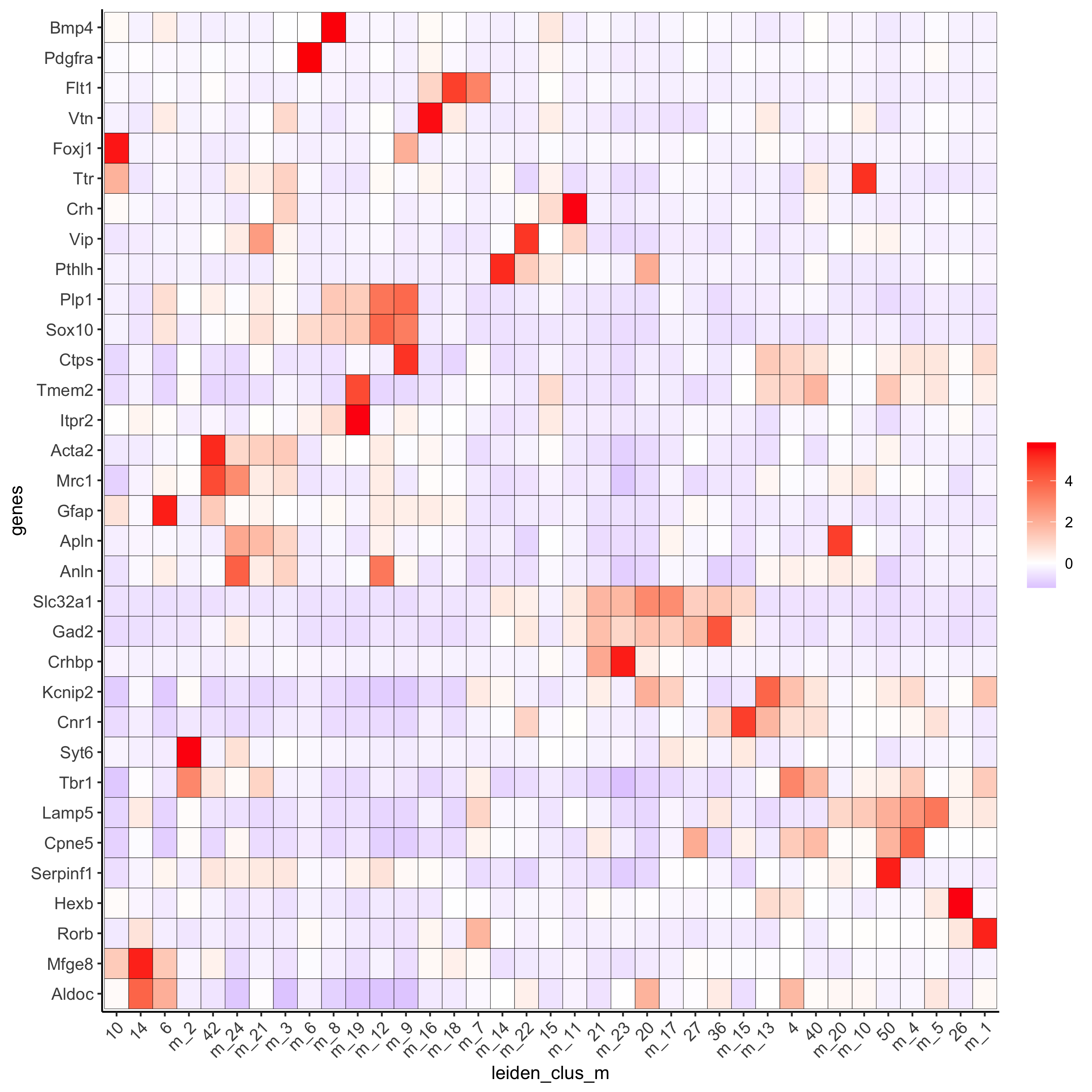
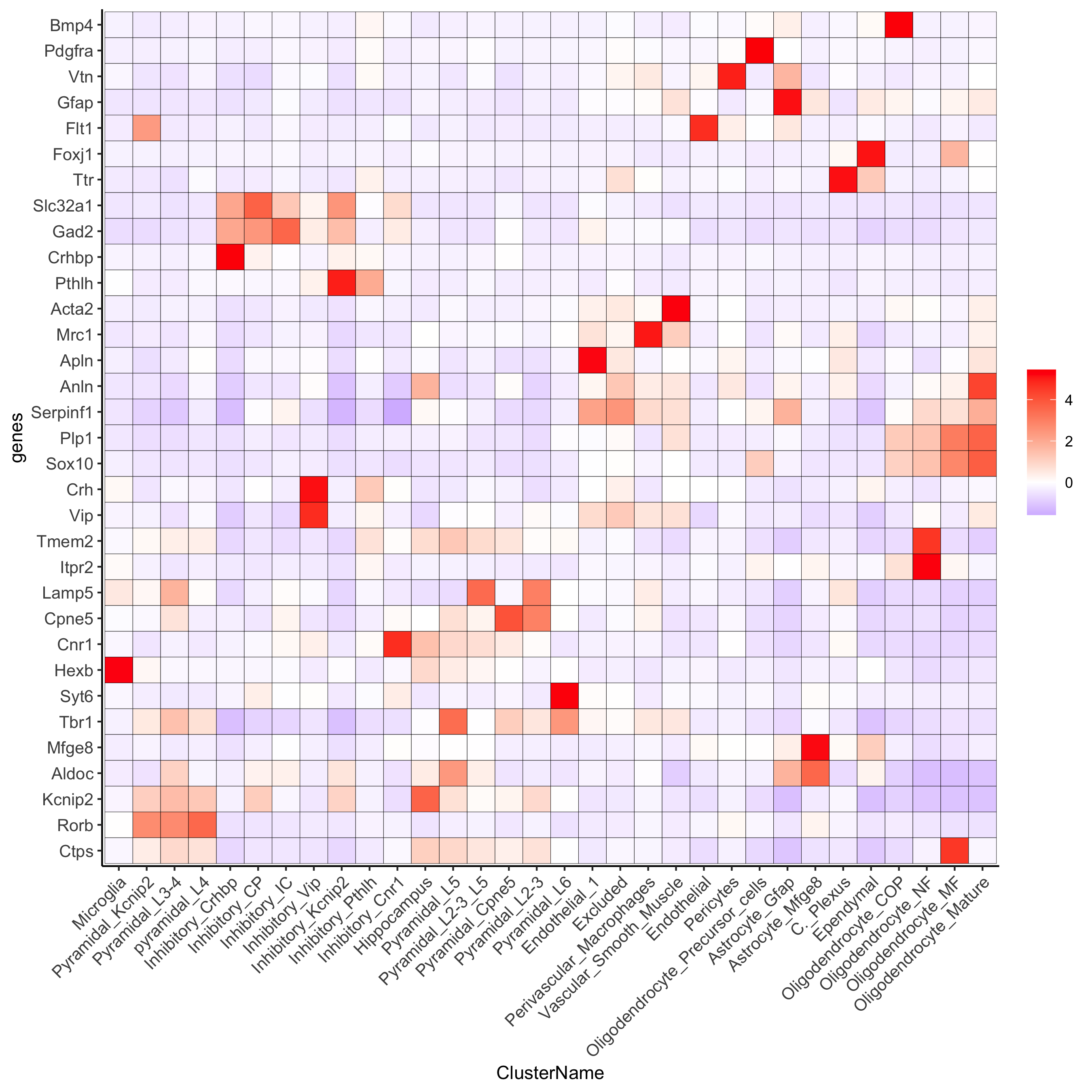
plotMetaDataHeatmap(osm_test, expression_values = 'custom',
metadata_cols = c('leiden_clus_m'),
save_param = c(save_name = '6_b_metaheatmap'))
plotMetaDataHeatmap(osm_test, expression_values = 'custom',
metadata_cols = c('leiden_clus_m'),
save_param = c(save_name = '6_e_metaheatmap_all_genes'))
plotMetaDataHeatmap(osm_test, expression_values = 'custom',
metadata_cols = c('ClusterName'),
save_param = c(save_name = '6_f_metaheatmap_all_genes_names'))
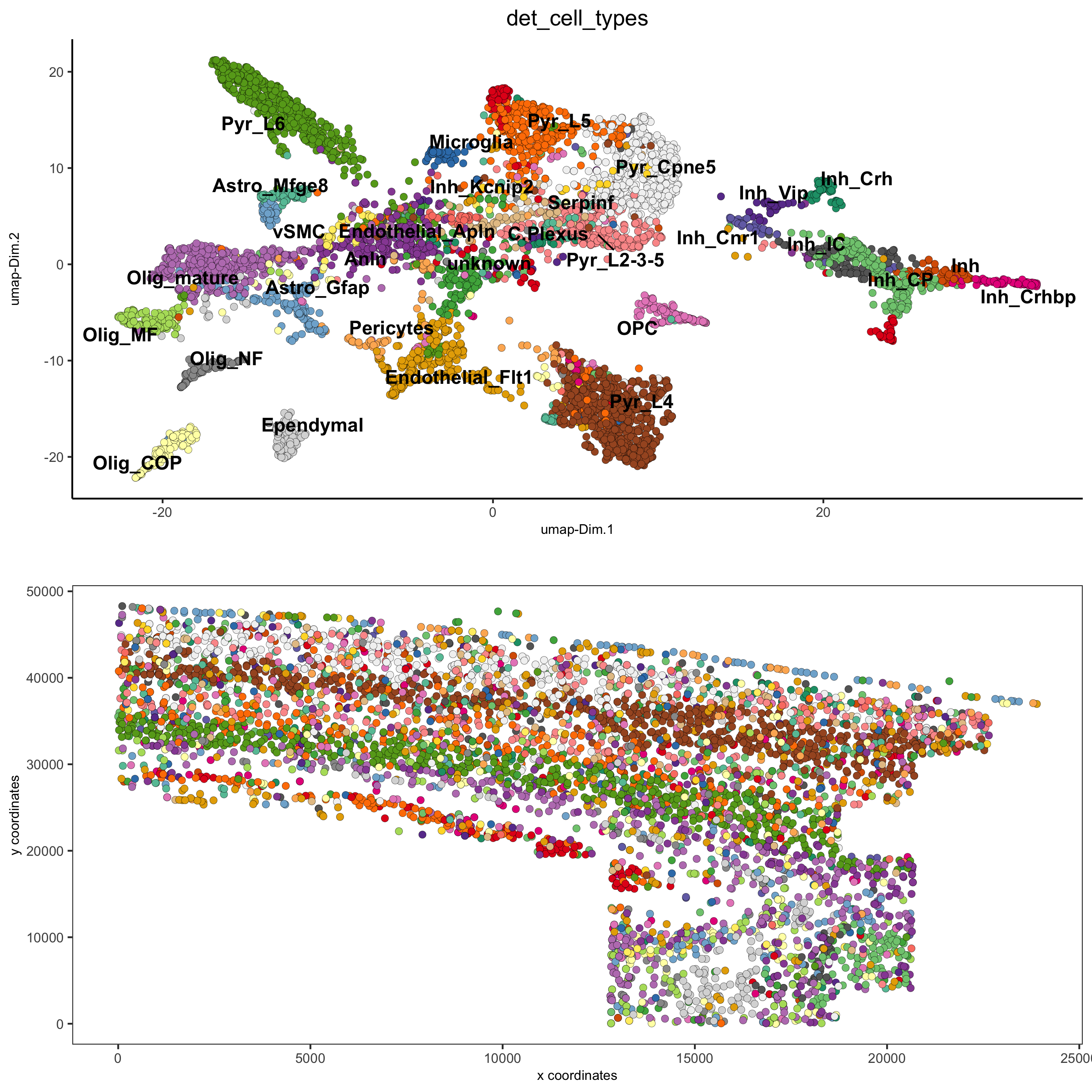
7. Cell-Type Annotation¶
## create vector with names
## compare clusters with osmFISH paper
clusters_det_SS_cortex = c('Ependymal', 'Astro_Mfge8', 'Astro_Gfap', 'Pyr_L6', 'vSMC',
'Anln', 'Anln', 'Anln', 'OPC', 'Olig_COP',
'Olig_NF', 'Olig_mature', 'Olig_MF', 'Pericytes', 'Endothelial_Flt1',
'Endothelial_Flt1', 'Inh_Kcnip2', 'Inh_Vip', 'unknown', 'Inh_Crh',
'Inh', 'Inh_Crhbp', 'Inh_CP','Inh_CP', 'Inh_IC',
'Inh_IC', 'Inh_Cnr1', 'Inh_Kcnip2', 'Pyr_L5', 'Pyr_L5',
'Endothelial_Apln', 'C.Plexus', 'Serpinf', 'Pyr_Cpne5', 'Pyr_L2-3-5',
'Microglia', 'Pyr_L4')
names(clusters_det_SS_cortex) = c('10', '14', '6', 'm_2', '42', 'm_24', 'm_21', 'm_3', 'm_6', 'm_8',
'm_19', 'm_12', 'm_9', 'm_16', 'm_18', 'm_7', 'm_14', 'm_22', '15', 'm_11',
'21', 'm_23', '20', 'm_17', '27', '36', 'm_15', 'm_13', '4', '40',
'm_20', 'm_10', '50', 'm_4', 'm_5', '26', 'm_1')
osm_test = annotateGiotto(gobject = osm_test, annotation_vector = clusters_det_SS_cortex,
cluster_column = 'leiden_clus_m', name = 'det_cell_types')
spatDimPlot2D(gobject = osm_test, cell_color = 'det_cell_types',dim_point_size = 2, spat_point_size = 2,
save_param = c(save_name = '7_a_annotation_leiden_merged_detailed'))
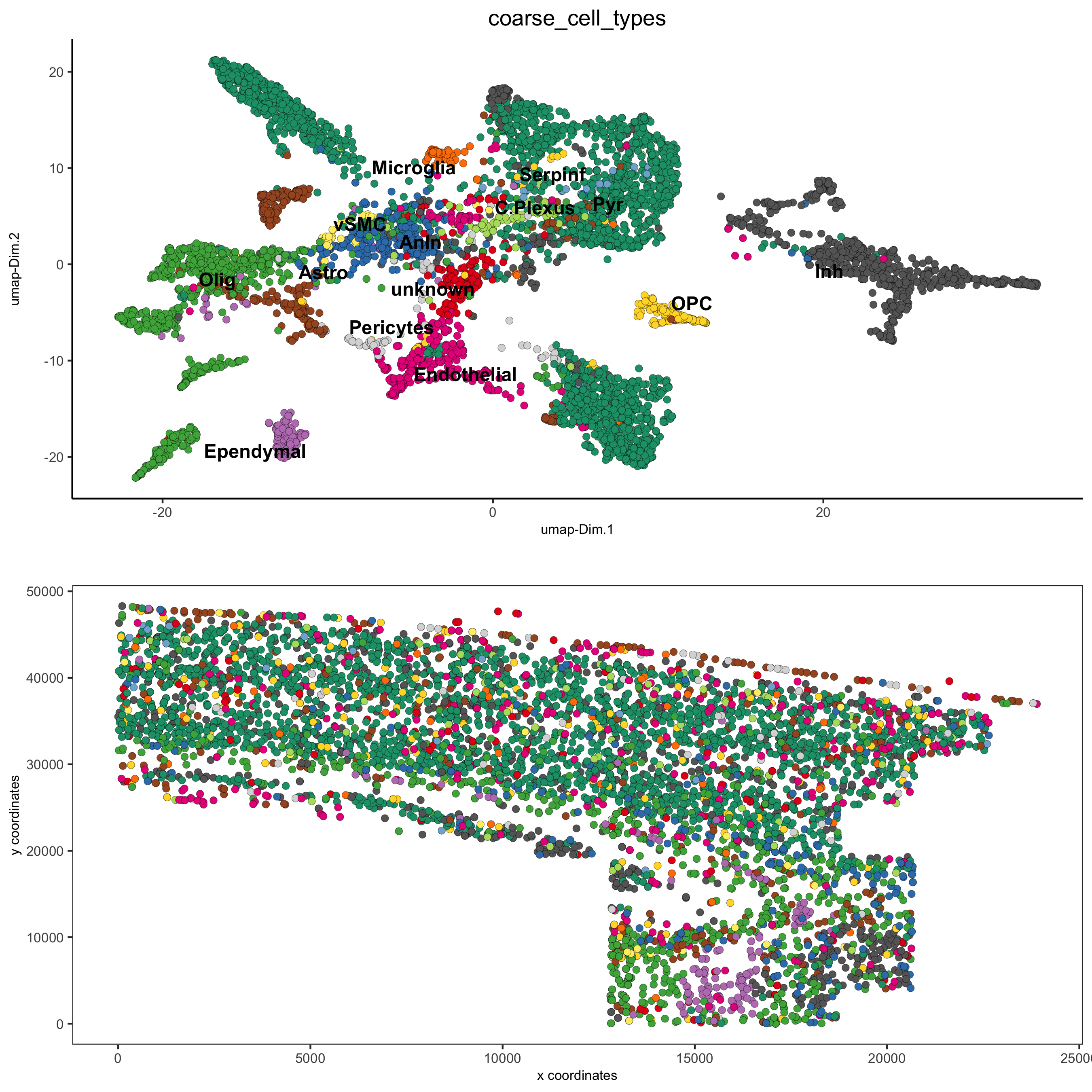
## coarse cell types
clusters_coarse_SS_cortex = c('Ependymal', 'Astro', 'Astro', 'Pyr', 'vSMC',
'Anln', 'Anln', 'Anln', 'OPC', 'Olig',
'Olig', 'Olig', 'Olig', 'Pericytes', 'Endothelial',
'Endothelial', 'Inh', 'Inh', 'unknown', 'Inh',
'Inh', 'Inh', 'Inh', 'Inh', 'Inh',
'Inh', 'Inh', 'Inh', 'Pyr', 'Pyr',
'Endothelial', 'C.Plexus', 'Serpinf', 'Pyr', 'Pyr',
'Microglia', 'Pyr')
names(clusters_coarse_SS_cortex) = c('Ependymal', 'Astro_Mfge8', 'Astro_Gfap', 'Pyr_L6', 'vSMC',
'Anln', 'Anln', 'Anln', 'OPC', 'Olig_COP',
'Olig_NF', 'Olig_mature', 'Olig_MF', 'Pericytes', 'Endothelial_Flt1',
'Endothelial_Flt1', 'Inh_Kcnip2', 'Inh_Vip', 'unknown', 'Inh_Crh',
'Inh', 'Inh_Crhbp', 'Inh_CP','Inh_CP', 'Inh_IC',
'Inh_IC', 'Inh_Cnr1', 'Inh_Kcnip2', 'Pyr_L5', 'Pyr_L5',
'Endothelial_Apln', 'C.Plexus', 'Serpinf', 'Pyr_Cpne5', 'Pyr_L2-3-5',
'Microglia', 'Pyr_L4')
osm_test = annotateGiotto(gobject = osm_test, annotation_vector = clusters_coarse_SS_cortex,
cluster_column = 'det_cell_types', name = 'coarse_cell_types')
spatDimPlot2D(gobject = osm_test, cell_color = 'coarse_cell_types',dim_point_size = 1.5, spat_point_size = 1.5,
save_param = c(save_name = '7_b_annotation_leiden_merged_coarse'))
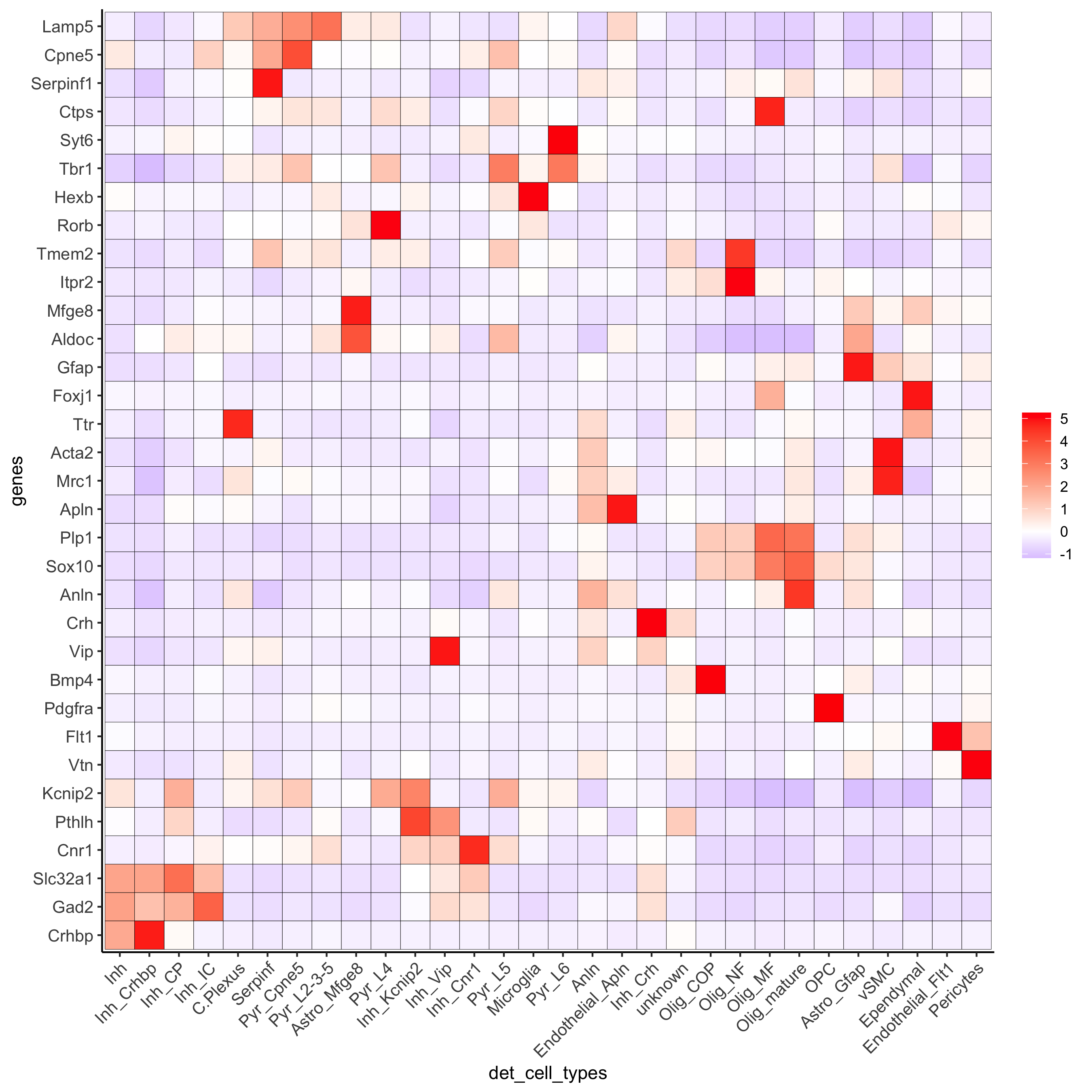
# heatmaps #
showClusterHeatmap(gobject = osm_test, cluster_column = 'det_cell_types',
save_param = c(save_name = '7_c_clusterHeatmap_det_cell_types', units = 'in'))

plotHeatmap(osm_test, genes = osm_test@gene_ID, cluster_column = 'det_cell_types',
legend_nrows = 2, expression_values = 'custom',
gene_order = 'correlation', cluster_order = 'correlation',
save_param = c(save_name = '7_d_heatamp_det_cell_types'))

plotMetaDataHeatmap(osm_test, expression_values = 'custom',
metadata_cols = c('det_cell_types'),
save_param = c(save_name = '7_e_metaheatmap'))
8. Spatial Grid¶
osm_test <- createSpatialGrid(gobject = osm_test,
sdimx_stepsize = 2000,
sdimy_stepsize = 2000,
minimum_padding = 0)
spatPlot2D(osm_test, cell_color = 'det_cell_types', show_grid = T,
grid_color = 'lightblue', spatial_grid_name = 'spatial_grid',
point_size = 1.5,
save_param = c(save_name = '8_grid_det_cell_types'))

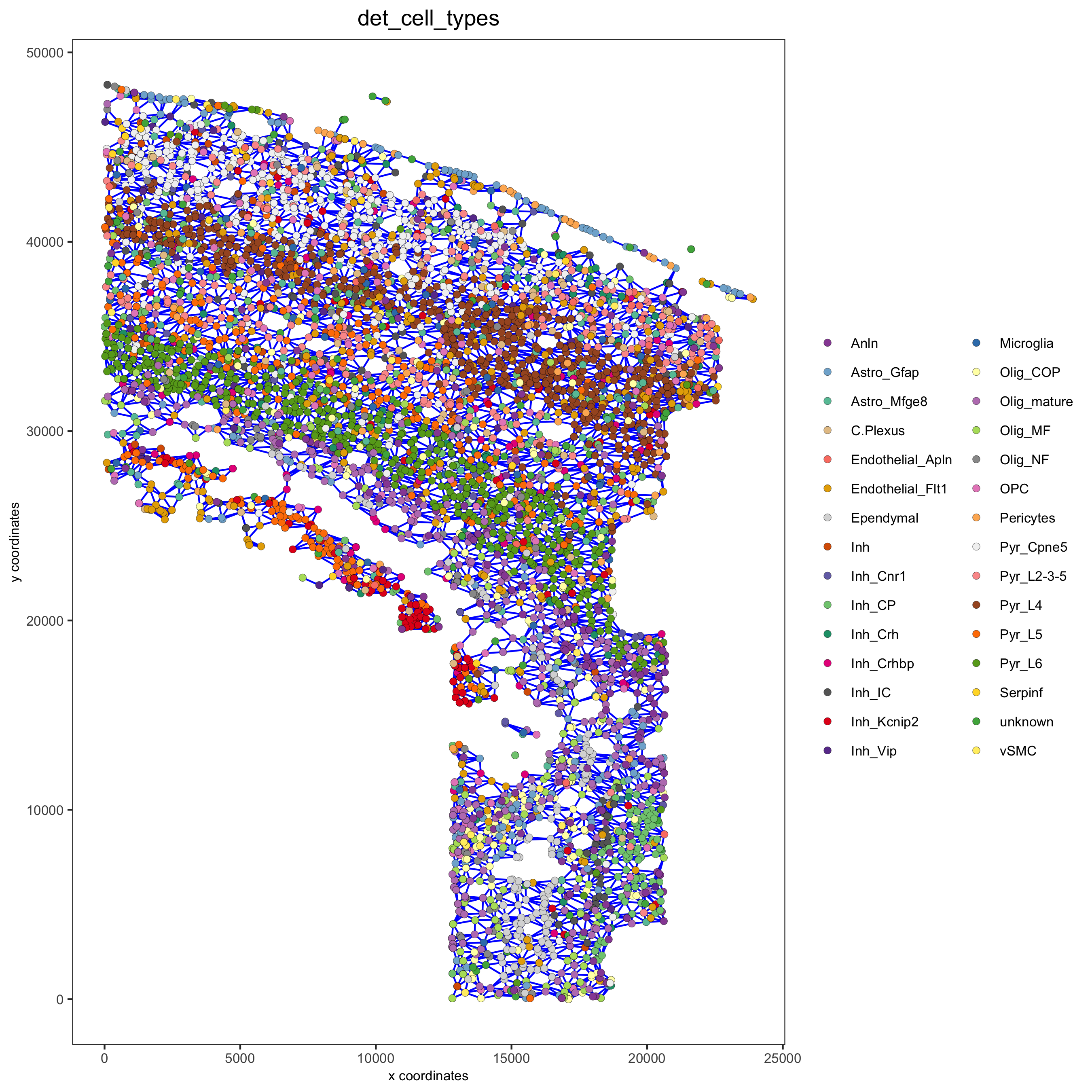
9. Spatial Network¶
osm_test <- createSpatialNetwork(gobject = osm_test)
spatPlot2D(gobject = osm_test, show_network = T,
network_color = 'blue',
point_size = 1.5, cell_color = 'det_cell_types', legend_symbol_size = 2,
save_param = c(save_name = '9_spatial_network_k10'))
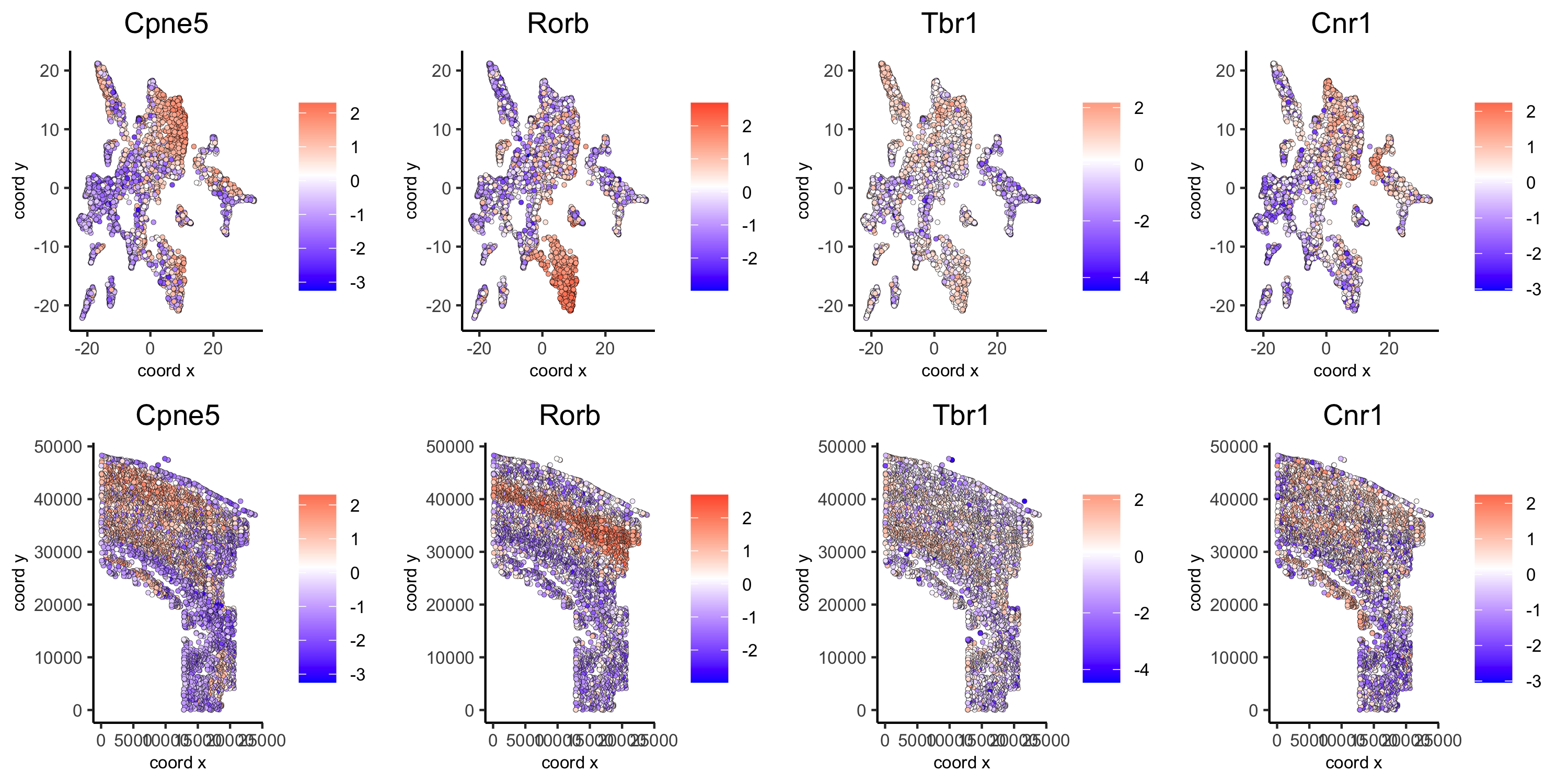
10. Spatial Genes¶
# km binarization
kmtest = binSpect(osm_test, bin_method = 'kmeans')
spatDimGenePlot2D(osm_test, expression_values = 'scaled',
genes = kmtest$genes[1:4],
plot_alignment = 'vertical', cow_n_col = 4,
save_param = c(save_name = '10_a_spatial_genes_km', base_height = 5, base_width = 10))
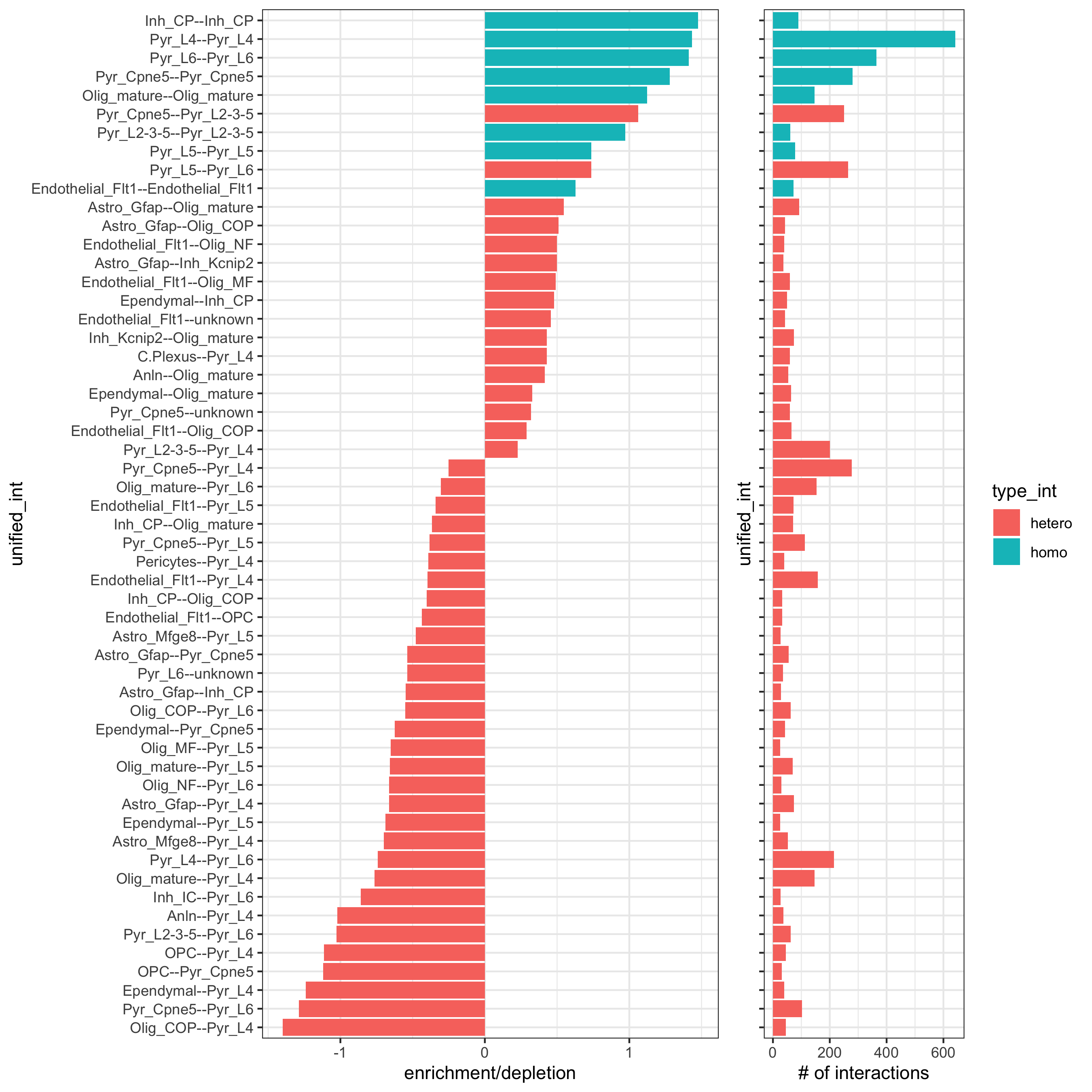
11. Cell-Cell Preferential Proximity¶
## calculate frequently seen proximities
cell_proximities = cellProximityEnrichment(gobject = osm_test,
cluster_column = 'det_cell_types',
number_of_simulations = 1000)
## barplot
cellProximityBarplot(gobject = osm_test, CPscore = cell_proximities, min_orig_ints = 25, min_sim_ints = 25,
save_param = c(save_name = '12_a_barplot_cell_cell_enrichment'))
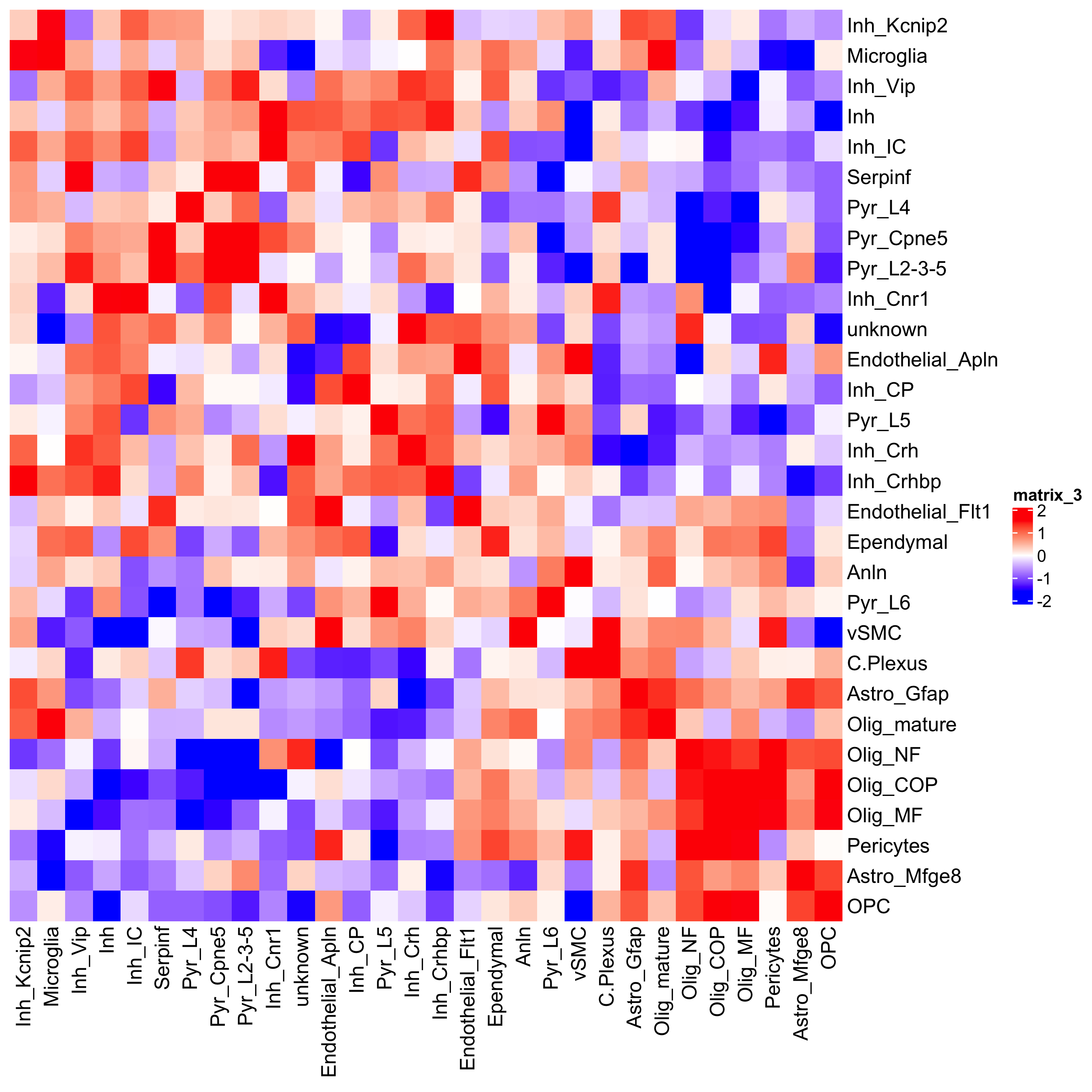
## heatmap
cellProximityHeatmap(gobject = osm_test, CPscore = cell_proximities, order_cell_types = T, scale = T,
color_breaks = c(-1.5, 0, 1.5), color_names = c('blue', 'white', 'red'),
save_param = c(save_name = '12_b_heatmap_cell_cell_enrichment', unit = 'in'))
## network
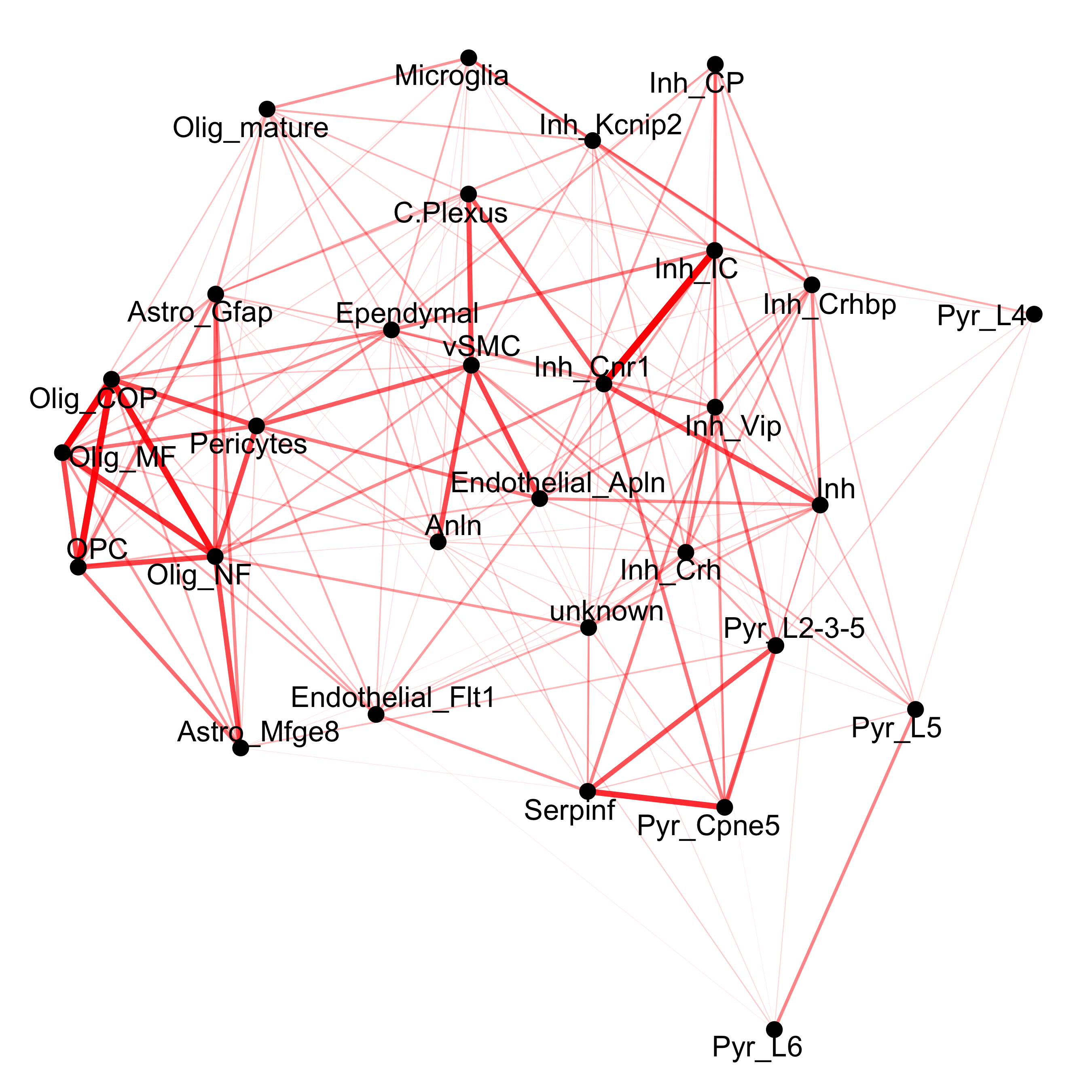
cellProximityNetwork(gobject = osm_test, CPscore = cell_proximities, remove_self_edges = T, only_show_enrichment_edges = T,
save_param = c(save_name = '12_c_network_cell_cell_enrichment'))
## visualization
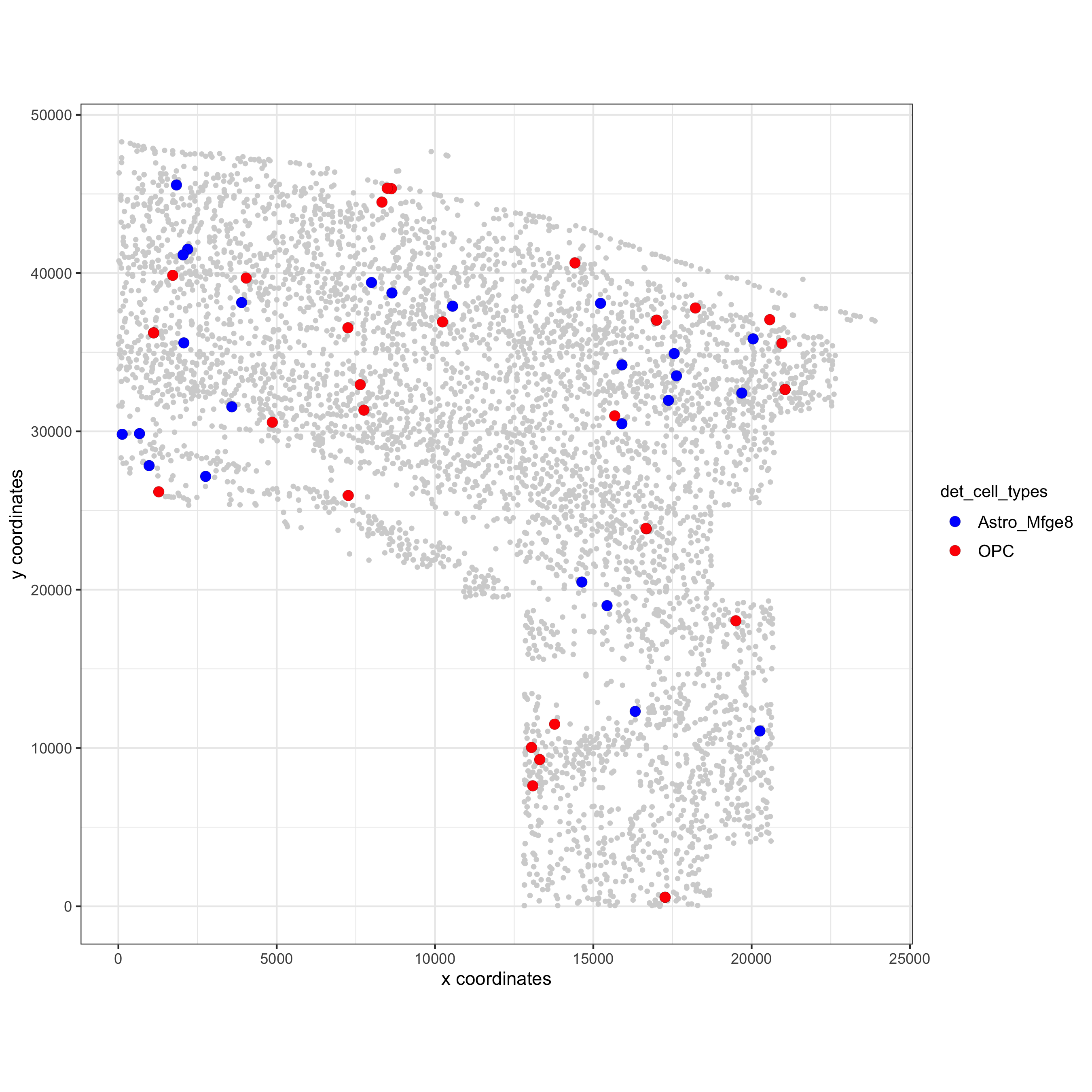
spec_interaction = "Astro_Mfge8--OPC"
cellProximitySpatPlot(gobject = osm_test,
interaction_name = spec_interaction,
cluster_column = 'det_cell_types',
cell_color = 'det_cell_types', cell_color_code = c('Astro_Mfge8' = 'blue', 'OPC' = 'red'),
coord_fix_ratio = 0.5, point_size_select = 3, point_size_other = 1.5,
save_param = c(save_name = '12_d_cell_cell_enrichment_selected'))