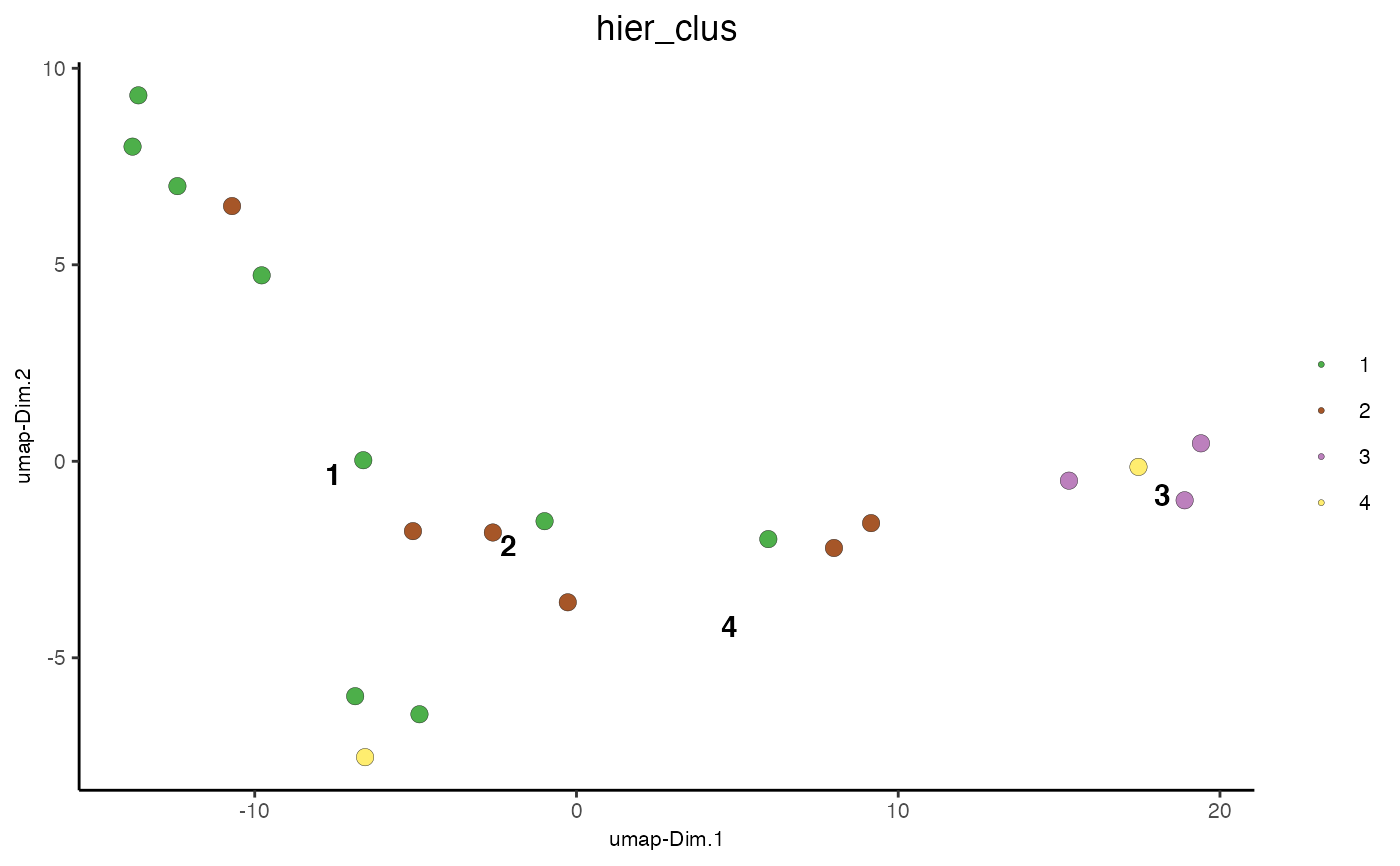
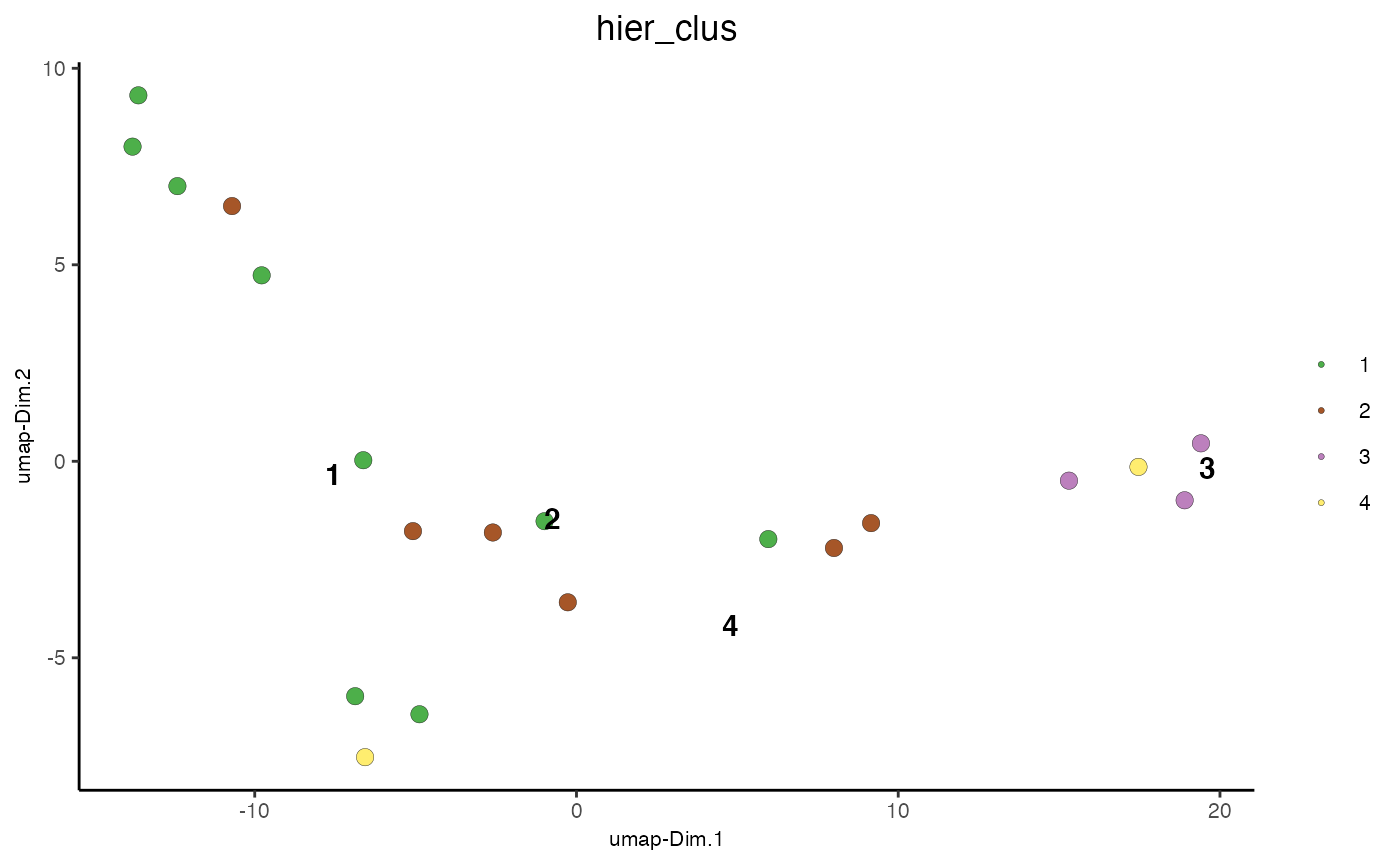
Cluster Cells Using Hierarchical Clustering¶
-
doHclust()
Cluster cells using hierarchical clustering algorithm.
ddoHclust(
gobject,
expression_values = c("normalized", "scaled", "custom"),
genes_to_use = NULL,
dim_reduction_to_use = c("cells", "pca", "umap", "tsne"),
dim_reduction_name = "pca",
dimensions_to_use = 1:10,
distance_method = c("pearson", "spearman", "original", "euclidean", "maximum",
"manhattan", "canberra", "binary", "minkowski"),
agglomeration_method = c("ward.D2", "ward.D", "single", "complete", "average",
"mcquitty", "median", "centroid"),
k = 10,
h = NULL,
name = "hclust",
return_gobject = TRUE,
set_seed = T,
seed_number = 1234
)
Arguments¶
gobject |
giotto object |
expression_values |
expression values to use |
genes_to_use |
subset of genes to use |
dim_reduction_to_use |
dimension reduction to use |
dim_reduction_name |
dimensions reduction name |
dimensions_to_use |
dimensions to use |
distance_method |
distance method |
agglomeration_method |
agglomeration method for hclust |
k |
number of final clusters |
h |
cut hierarchical tree at height = h |
name |
name for hierarchical clustering |
return_gobject |
boolean: return giotto object (default = TRUE) |
set_seed |
set seed |
seed_number |
number for seed |
Value¶
Giotto object with new clusters appended to cell metadata
Details¶
Description on how to use hierarchical clustering method.
Examples¶
data(mini_giotto_single_cell)
mini_giotto_single_cell = doHclust(mini_giotto_single_cell, k = 4, name = 'hier_clus')
plotUMAP_2D(mini_giotto_single_cell, cell_color = 'hier_clus', point_size = 3)