Identify Significant PCs With A Screeplot¶
-
screePlot()
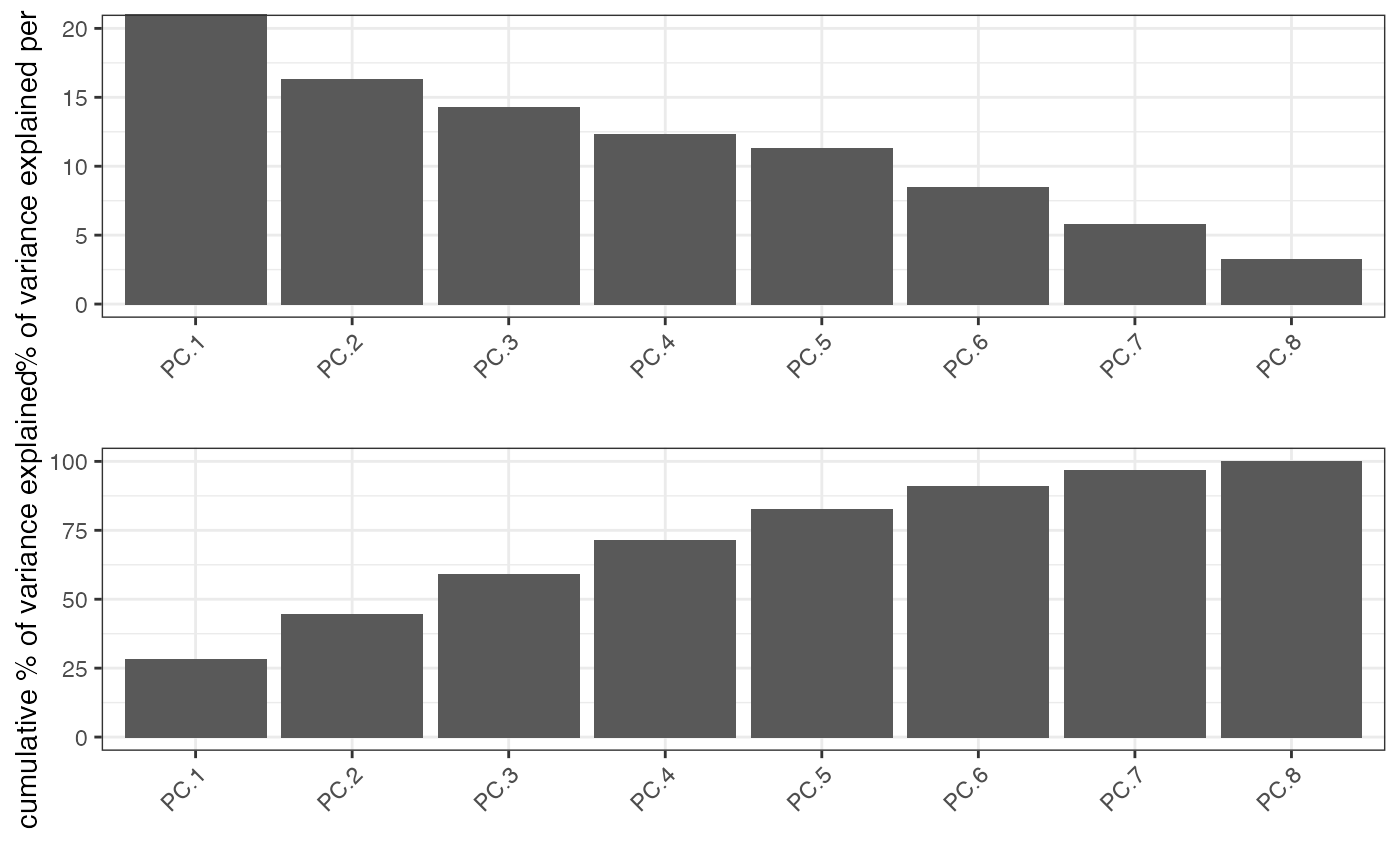
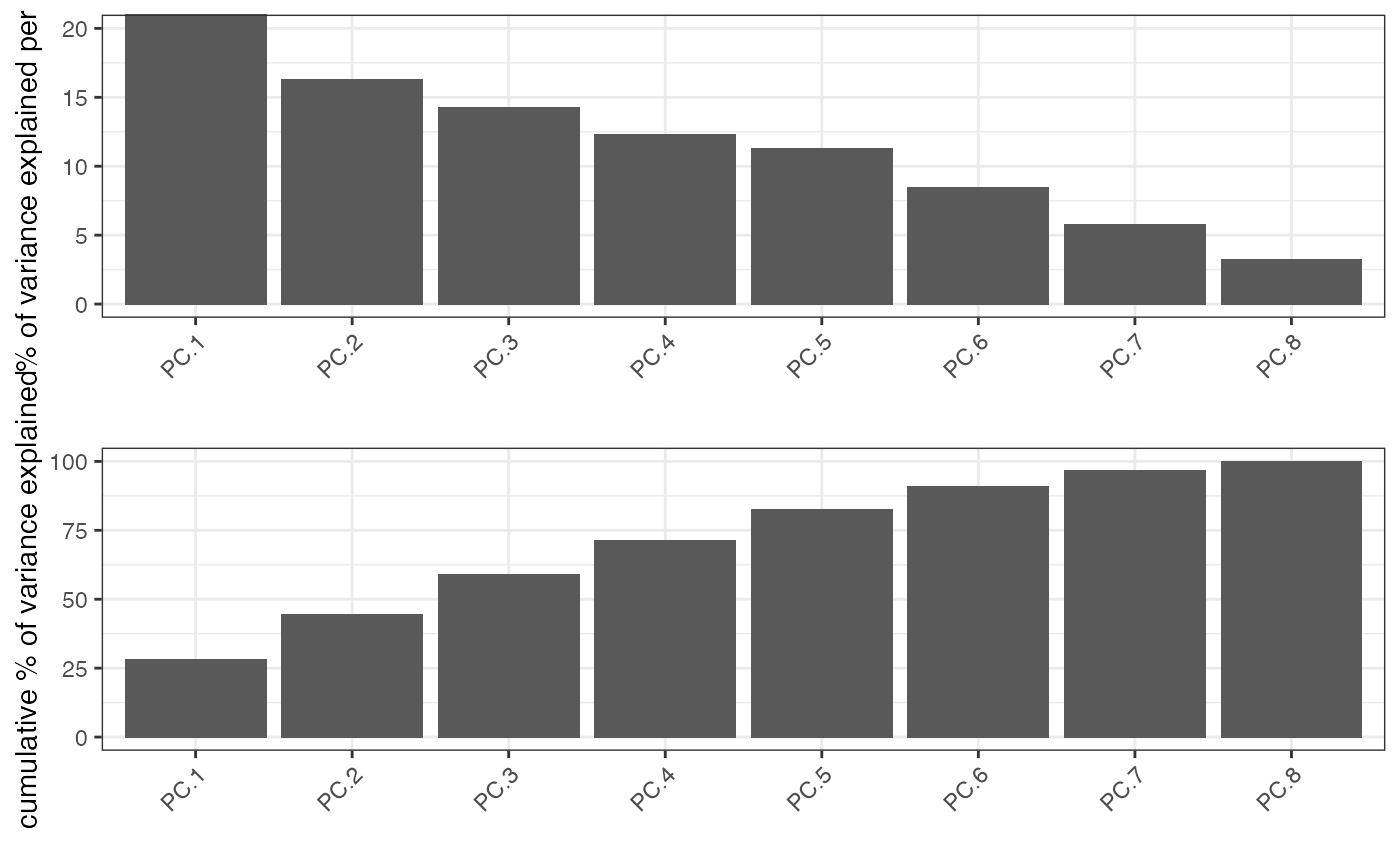
Identify significant prinicipal components (PCs) using a screeplot (a.k.a. elbow-plot).
screePlot(
gobject,
name = "pca",
expression_values = c("normalized", "scaled", "custom"),
reduction = c("cells", "genes"),
method = c("irlba", "factominer"),
rev = FALSE,
genes_to_use = NULL,
center = F,
scale_unit = F,
ncp = 100,
ylim = c(0, 20),
verbose = T,
show_plot = NA,
return_plot = NA,
save_plot = NA,
save_param = list(),
default_save_name = "screePlot",
...
)
Arguments¶
gobject |
giotto object |
name |
name of PCA object if available |
expression_values |
expression values to use |
reduction |
cells or genes |
method |
which implementation to use |
rev |
do a reverse PCA |
genes_to_use |
subset of genes to use for PCA |
center |
center data before PCA |
scale_unit |
scale features before PCA |
ncp |
number of principal components to calculate |
ylim |
y-axis limits on scree plot |
verbose |
verobsity |
show_plot |
show plot |
return_plot |
return ggplot object |
save_plot |
directly save the plot [boolean] |
save_param |
list of saving parameters from all_plots_save_function() |
default_save_name |
default save name for saving, don’t change, change save_name in save_param |
… |
additional arguments to pca function, see runPCA |
Value¶
ggplot object for scree method.
Details¶
Screeplot works by plotting the explained variance of each individual PC in a barplot allowing you to identify which PC provides a significant contribution (a.k.a ‘elbow method’). Screeplot will use an available pca object, based on the parameter ‘name’, or it will create it if it’s not available (see runPCA()).
Examples¶
data(mini_giotto_single_cell)
screePlot(mini_giotto_single_cell, ncp = 10)
#> PCA with name: pca already exists and will be used for the screeplot